Our Bodies Are Formed Streams
“The method of nature: who could ever analyse it? That rushing stream will not stop to be observed. We can never surprise nature in a corner; never find the end of a thread; never tell where to set the first stone. The bird hastens to lay her egg: the egg hastens to be a bird … [The world’s] smoothness is the smoothness of the pitch of the cataract. Its permanence is a perpetual inchoation. Every natural fact is an emanation, and that from which it emanates is an emanation also, and from every emanation is a new emanation. If anything could stand still, it would be crushed and dissipated by the torrent it resisted ...”
In this materialist era, we like our reality hard and our truths weighty and rock solid. We may accept that there are states of matter less substantial than rocks, but in our imaginations we turn even fluids and gases into collections of tiny particles. Similarly, in our reconstructions of physiological processes, material structures come first, and only then can movement, flow, and meaningful activity somehow occur.
How, after all, can there be movement without things to do the moving? (It’s easy to forget that energy, fields, and forces are not things!) Ask someone to describe the circulatory system, and you will very likely hear a great deal about the heart, arteries, veins, capillaries, red blood cells, and all the rest, but little or nothing about the endless subtleties of circulatory movement. And yet, embryological development shows that
the body does not behave like a plumber, first connecting the water pipes in a house and then turning the water on … the first blood-like liquid … simply trickles through gaps in the tissues … Preferred channels develop only very gradually as blood cells are deposited along the edges and eventually merge into the beginnings of vessel walls (Schad 2002, p. 80).
The situation loosely reminds one of college campuses when new lawn is laid down. Landscapers typically wait to see where human traffic creates clear pathways through the grass before “solidifying” the paths with concrete.
Moreover, “when blood vessels first start to form, the heart does not yet exist … early blood flow stimulates the development of the heart” (Schad 2002, pp. 82-83). Again, form arises from movement. Thus, the spiraling fibers of the heart muscle that help to direct the blood in its flow are themselves a congealed image of the swirling vortex of blood within. This kind of mutuality holds even for the heart’s basic structural divisions:
Before the heart has developed walls (septa) separating the four chambers from each other, the blood already flows in two distinct “currents” through the heart. The blood flowing through the right and left sides of the heart do not mix, but stream and loop by each other, just as two currents in a body of water. In the “still water zone” between the two currents, the septum dividing the two chambers forms. Thus the movement of the blood gives the parameters for the inner differentiation of the heart, just as the looping heart redirects the flow of blood 1 (Holdrege 2002, p. 12).
There is no escaping the fact that we begin our lives in a thoroughly fluid and plastic condition. Only with time do relatively solid and enduring structures precipitate out as tentatively formed “islands” within the streaming rivers of cells that shape the life of the early embryo. Movement gives rise to structures, structures do not give rise to movement. As adults, we are still about seventy percent water.
One might think quite differently based on the scientific rhetoric to which we are daily exposed. This could easily lead us to believe that the real essence and solid foundation of our lives was from the beginning rigidly established inside those very first cells. There we find DNA macromolecules that, in a ceaseless flood of images, are presented to us as crystalline forms in the shape of a spiraling ladder — a ladder whose countless rungs constitute the fateful stairway of our lives. So, too, with the proteins and protein complexes of our bodies: we have been told for decades that they fold precisely into wondrously efficient molecular machines whose all-important functions are predestined by the DNA sequence.
The trouble is, biological researches of the last few decades have not merely hinted at an altogether different story; they have (albeit sometimes to deaf ears) been trumpeting it aloud as a theme with a thousand variations. Even the supposedly “solid” structures and molecular complexes in our cells — including the ones we have imagined as strict determinants of our lives — are caught up in functionally significant movement that the structures themselves can hardly have originated. (See Chapter 3, “What Brings Our Genome Alive?” and Chapter 4, “The Sensitive, Dynamic Cell.”)
Nowhere are we looking either at a static sculpture or at controlling molecules responsible for the sculpting. In an article in Nature following the completion of the Human Genome Project, Helen Pearson (2003) interviewed many geneticists in order to assemble the emerging picture of DNA. One research group, she reported, has shown that the molecule is made “to gyrate like a demonic dancer.” Others point out how chromosomes “form fleeting liaisons with proteins, jiggle around impatiently and shoot out exploratory arms.” Phrases such as “endless acrobatics,” “subcellular waltz,” and DNA that “twirls in time and space” are strewn through the article. “The word ‘static’ is disappearing from our vocabulary,” remarks cell biologist and geneticist Tom Misteli, a Distinguished Investigator at the National Cancer Institute in Bethesda, Maryland.
Everywhere we look, shifting form and movement show themselves to be the “substance” of biological activity. The physiological narratives of our lives play out in gestural dramas that explain the origin and significance of structures rather than being explained by those structures.
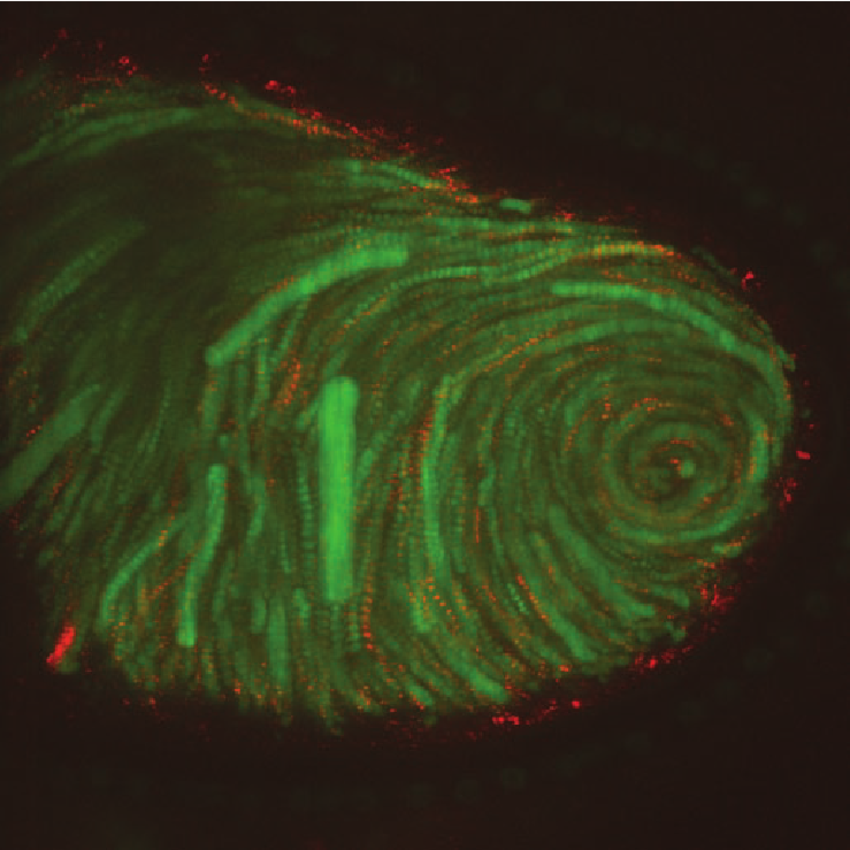
Figure 5.1. Multiple, superimposed images from a movie, showing movements in a fruit fly oocyte (a developing egg). Yolk granules are stained green, and tiny red fluorescent polystyrene beads have been injected into the egg to show the dynamism of flow in the egg body over time.2
Hannah Landecker, a professor of both genetics and sociology at UCLA, having looked at the impact of recent, highly sophisticated cellular imaging techniques on our understanding, has written: “The depicted cell seems a kind of endlessly dynamic molecular sea, where even those ‘structures’ elaborated by a century of biochemical analysis are constantly being broken down and resynthesized.” And she adds: “It is not so much that the structures begin to move, but movements — for example in the assembly and self-organization of the cytoskeleton — begin to constitute structure” (Landecker 2012). See Figure 5.1.
And a team of biochemists from Duke and Stanford Universities point out how inadequate is our knowledge of the action of biomolecules when all we have is a frozen structure of the sort commonly reported in the literature. “In reality,” they say, “all macromolecules dynamically alternate between conformational states [that is, between three-dimensional folded shapes] to carry out their biological functions”:
Decades ago, it was realized that the structures of biomolecules are better described as “screaming and kicking,” constantly undergoing motions on timescales spanning twelve orders of magnitude, from picoseconds [trillionths of a second] to seconds (Ganser et al. 2019).
Why, after all, should we ever have expected our physiology to be less a matter of gesturings than is our life as a whole?
A long way from
crystalline order
According to the old story of the machine-organism, a protein-coding DNA sequence, or gene, is not only mirrored in an exact messenger RNA (mRNA) sequence, but the mRNA in turn is translated into an exact amino acid sequence in the resulting protein, which finally folds into a fixed shape predestined by that sequence. It was a picture of perfect, lawful, lockstep necessity, leading from DNA through mRNA to a final, functional protein.
“There is a sense,” wrote Richard Dawkins, “in which the three-dimensional coiled shape of a protein is determined by the one-dimensional sequence of code symbols in the DNA.” Further, “the whole translation, from strictly sequential DNA read-only memory to precisely invariant three-dimensional protein shape, is a remarkable feat of digital information technology” (Dawkins 2006, p. 171).
And these proteins in turn were thought to carry out their functions by neatly engaging with each other in a machine-like manner, snapping into place like perfectly matched puzzle pieces or inserting into each other like keys in locks.
We now know, and already knew when Dawkins published those words, that everything about this narrative was wrong — and not only the parts about DNA and RNA. Among proteins (those “workhorses of the cell”) every individual molecule lives in transformational movement — as a dynamic ensemble of rapidly “morphing,” or interconverting, conformations — and therefore does not have a “precisely invariant three-dimensional shape.”
But there is much more that wholly escaped Dawkins’ computerized imagination. Quite apart from the fact that each protein molecule rapidly shifts between distinctly different, folded structures, we now know that intrinsically disordered proteins — proteins that, in whole or in part, have no particular, inherent structure at all — are crucial for much of a cell’s functioning. Researchers refer to “fluid-like” and “surface-molten” proteins (Grant et al. 2010; Zhou et al. 1999). This is why biophysicist Konstantin Turoverov and his Russian and American colleagues tell us that “the model of the organization of living matter is changing to one described by highly dynamic biological soft matter.” For decades, they note, protein interactions were “considered to be rigid, where, for a given protein, a unique 3D structure defined a unique biological activity.” However,
it is now realized that many protein functions rely on the lack of specific structure. This recognition has changed the classical consideration of a functioning protein from a quasi-rigid entity with a unique 3D structure resembling an aperiodic crystal into a softened conformational ensemble representation, with intrinsic disorder affecting different parts of a protein to different degrees3 (Turoverov et al. 2019, emphasis added).
Clearly, the finally achieved protein need not be anything like the predetermined, inflexible mechanism with a single, well-defined structure imagined by Dawkins. Proteins can be true shape-shifters, responding and adapting to an ever-varying context — so much so that (as the noted experimental cell biologist, Stephen Rothman, has written) the “same” proteins with the same amino acid sequences can, in different environments, “be viewed as totally different molecules” with distinct physical and chemical properties (Rothman 2002, p. 265).
Many intrinsically unstructured proteins are involved in regulatory processes, and often serve as Proteus-like hub elements at the center of large protein interaction networks (Gsponer and Babu 2009). They also play a decisive role in molecular-level communication within and between cells, where their flexibility allows them to modulate or even reverse the typical significance of a signal,4 in effect transforming do this into don’t do this or do that (Hilser 2013).
But the troubling question arises: if unstructured proteins, or unstructured regions in proteins, are not “pre-fitted” for particular interactions — if, in their “molten” state, they have boundless possibilities for interacting with other molecules and even for reversing the effects of those other molecules — how do these proteins “know” what to do at any one place and time (Talbott 2024)? Or, as one pair of researchers put it, “How is the logic of molecular specificity encoded in the promiscuous interactions of intrinsically disordered proteins?” (Zhu and Brangwynne 2015). In a following section (“The unexpected phases of life”) we will look at one of the most recent and dramatic developments in cellular physiology, which has seemed to many biologists to offer an approach to this problem.
But first we should note the continuing mechanistic bias in the negative descriptors, “disordered” and “unstructured,” which I have grudgingly adopted from the conventional literature. Contrary to this usage, the loose, shifting structure of a protein need be no more disordered than the graceful, swirling currents of a river or the movements of a ballet dancer. Given the many living processes these proteins harmoniously support and participate in (including, in fact, the movements of the ballet dancer), it would be strange to assume that their performance is anything less than graceful, artistic, purposive, and meaningful.
Fluid, “living” molecules do not lend themselves to the analogy with mechanisms, which may explain why the mistaken idea of precisely articulated, folded parts was so persistent, and why the recognition of unstructured proteins was so late in coming. Indeed, this recognition has only recently been dawning upon the biological community as a whole, a fact that led to this lament as late as 2008 at a conference on “bioinformatics and bioengineering” at Harvard Medical School:
Experimentalists have been providing evidence over many decades that some proteins lack fixed structure or are disordered (or unfolded) under physiological conditions. In addition, experimentalists are also showing that, for many proteins, their functions depend on the unstructured rather than structured state; such results are in marked contrast to the greater than hundred year old views such as the lock and key hypothesis. Despite extensive data on many important examples, including disease-associated proteins, the importance of disorder for protein function has been largely ignored. Indeed, to our knowledge, current biochemistry books don’t present even one acknowledged example of a disorder-dependent function, even though some reports of disorder-dependent functions are more than fifty years old (Dunker et al. 2008).
The unexpected
phases of life
It has become increasingly clear in recent years that, quite apart from its cytoskeleton and membrane-bound organelles (Chapter 4, “The Sensitive, Dynamic Cell”), the fluid cytoplasm in each cell is elaborately and “invisibly” organized. Various macromolecular complexes and other molecules, in more or less defined mixes, congregate in specific locations and sustain a collective identity, despite being unbounded by any sort of membrane. Here we’re looking at significant structure, or organization, without even a pretense of mechanically rigid form. How do cells manage that?
The problem was framed this way by Anthony Hyman from the Max Planck Institute of Molecular Cell Biology and Genetics in Dresden, Germany, and Clifford Brangwynne from the Department of Chemical and Biological Engineering at Princeton University:
Non-membrane-bound macromolecular assemblies found throughout the cytoplasm and nucleoplasm … consist of large numbers of interacting macromolecular complexes and act as reaction centers or storage compartments … We have little idea how these compartments are organized. What are the rules that ensure that defined sets of proteins cluster in the same place in the cytoplasm?
Even more puzzling, a “compartment” can maintain its functional (purposive) identity despite the rapid exchange of its contents with the surrounding cytoplasm. “Fast turnover rates of complexes in compartments can be found throughout the cell. How do these remain as coherent structures when their components completely turn over so quickly?” (Hyman and Brangwynne 2011).
Well-structured droplets
Part of the picture that has recently come into focus has to do with the phases of matter and the transitions between these phases. (Think, for example, of the solid, liquid, and gaseous phases of water, or of solutions and gels — matter in different states.) For example, it’s possible for well-defined droplets of one kind of liquid to occur within a different liquid, like oil droplets in water.
We now know that molecular complexes containing both RNA and protein often gather together to form distinctive RNA-protein liquids that separate out as droplets within the larger cytoplasmic medium. Like liquids in general, these droplets tend toward a round shape, can coalesce or divide, can wet surfaces such as membranes, and can flow. The concentration of particular molecules may be much greater in the droplets than in the surrounding fluid, conferring specific and efficient functions upon the assemblies.
Enzymes and reactants can rapidly diffuse within the liquid droplet, while also moving with relative ease across the boundary between droplet and surrounding medium. Yet this boundary can remain distinct until phase-changing environmental conditions occur — conditions that might involve slight changes in temperature, pH, salt concentration, electrical charge, molecular densities, the addition of small chemical groups to proteins, degradation of proteins, the activity of gene transcription, or still other factors.
In this way, a very subtle change — originating, say, from an extracellular influence — can yield a dramatic transformation of cytoplasmic organization, just as a slight change in the temperature or salinity of water can shift an ice-forming condition to an ice-melting one, or vice versa.
Box 5.1
On Shape-Shifting Blobs
Here are a few comments from an article in Nature titled “The Shape-Shifting Blobs That Rule Biology” (Dolgin 2022):
“For years, if you asked a scientist how they pictured the inner workings of a cell, they might have spoken of a highly organized factory, with different departments each performing specialized tasks in delineated assembly lines.
“Ask now, and they might be more inclined to compare the cell to a chaotic open-plan office, with hot-desking zones where different types of cellular matter gather to complete a task and then scatter to other regions.
“Everywhere scientists look in cells, throngs of proteins and RNA seem to be sticking together, coalescing into pearl-like droplets distinct from their surrounding environment. These dynamic compartments allow cells to perform essential functions, ranging from gene control and DNA repair to waste disposal and stress responses. They are often fleeting, and are unhindered by an enclosing membrane — unlike many other cellular components, such as mitochondria, which are membrane-bound. When a droplet is no longer needed, it vanishes.”
“One particular scaffolding protein seems to be the epicentre of stress-granule assembly. When the cell encounters adversity, this protein, called G3BP1, changes shape, prompting nearby RNA molecules to link up with it and promote clustering.”
“A catch-all name for these compartments: biomolecular condensates. The name left open how these assemblages of proteins and nucleic acids took shape or became undone. ‘It was deliberately supposed to be mechanism-free’ [explained one biologist] … Further experiments and theory showed that a huge number of forces work together to create condensates.”
“‘There isn’t a cellular process that’s been studied that is not now known to involve condensates’ [biologist Rick] Young says. ‘It involves damn near everything’.”
Moreover, these phase-separated droplets can be highly organized internally: “multiple distinct liquid phases can coexist and give rise to richly structured droplet architectures determined by the relative liquid surface tensions” (Shin and Brangwynne 2017). Also, some droplets may become gel-like,5 while others may form more or less solid granules. Many such droplets may pass through stages, from more liquid to more solid, before dispersing. They form in response to particular needs, perform their work, and then pass away. Others are more or less permanent. Phase separation has been called “a fundamental mechanism for organizing intracellular space” (Shin and Brangwynne 2017) — one where “function derives not from the structures of individual proteins, but instead, from dynamic material properties of entire [protein aggregates] acting in unison through phase changes” (Halfmann 2016).
We also know now that weak, transient interactions among intrinsically unstructured proteins and RNAs can result in crucial, flexible “scaffolds” that help to assemble these phase-separated aggregates, drawing in a set of functionally related molecules. “Weak,” “transient,” and “flexible” in my description here might be taken as indicators of the living, responsive, and non-machine-like character of the activity.
When things happen in the cell, phase transitions often play decisive roles, as a University of Colorado group discovered when looking at phase transitions in a roundworm. According to the researchers, these transitions “are controlled with surprising precision in early development, leading to starkly different supramolecular states” with altered organization and dynamics. “Reversible interactions among thousands of [these phase-separated] complexes,” the authors found, account for “large-scale organization of gene expression pathways in the cytoplasm” (Hubstenberger et al. 2013).
How do you regulate flow and phases?
All this is, if you think about it, an amazing departure from the kind of picture once burned into the minds of biologists such as Richard Dawkins, from whom we heard some errant words above. Once there were dreams of compelling digital instructions in DNA; of machine-like interactions between molecules; of deterministic formation and functioning of proteins; of the cell as a collection of distinct, well-defined structures; and of cellular processes with fully predictable outcomes. But this dream has faded in the clear daylight of an entirely different reality where, among many other things, we watch a subtle and almost incomprehensible play of material changes of state.
These state changes can be affected by infinitely varying factors, such as the momentary interaction between a few molecules of a particular sort, the “minor” modification of a molecule, the increasing concentration of molecules in a particular location, or the slight temperature change of a degree or two — the kind of change that, in the larger world of nature, can freeze the surface of a lake where, a few days previously, fish routinely breached the surface to feed on insects.
Ice cools a drink, water carves a canyon, steam powers a locomotive … But ice brings down power lines, water floods towns, steam scalds skin. The context for these states matters, and there can be consequences if the appropriate state is perturbed or dysregulated. Now more than ever, we understand that physical states dictate biological function, and … recent papers have highlighted, at the subcellular and tissue levels, the importance of understanding those states and the conditions in which they occur. (Szewczak 2019)
We heard it asked earlier how intrinsically unstructured proteins “know” what to do at any one place and time. The old model assumed, rather puzzlingly, that random encounters between freely diffusing molecules accounted for many of the biological interactions we observe. But numerous researchers are now embracing the emerging picture of biological phase transitions as offering a very different understanding. Peter Tompa, a structural biologist from Vrije Universiteit Brussel in Belgium, sees certain phase transitions as directing “the movement of regulatory proteins in and out of organized subcellular domains” — part of the systematic maintenance of order in the cell6 (Tompa 2013).
This is all well and good, but does it tell us (as is often implied) what “controls” and “directs” molecular engagements in relation to the distinct needs of the cell at different locations and times? If the organization of phase-separated aggregates is what coordinates the activity of proteins, then we shouldn’t have to ask, as researchers are now asking, “Why do some proteins localize to only the nucleolus, while others can be found in both the nucleolus and Cajal bodies?” (Zhu and Brangwynne 2015). (Cajal bodies, like the nucleolus, are non-membrane-bound organelles found in the cell nucleus.) And, even if that question had a ready answer, the more fundamental issue would remain: if we assume that phase-separated droplets lead to properly coordinated protein interactions, then what explains the well-timed and intricately organized formation, structuring, and dissolution of the condensates?
This illustrates how (to get ahead of ourselves just a little bit) all attempts to answer questions of regulation in strictly physical terms never do really answer them. Rather, they lead only to an elucidation of previous physical states that again raise the same broad questions. There is no way to step outside the endlessly regressing physical explanations except by truly stepping outside them — except, that is, by turning to a different sort of explanation possessing a certain “finalistic” aspect. This is where we attend to the play of intentions and end-directed activities that are implicit in the stories we find ourselves looking at.
After all, questions about biological regulation are questions about the significant patterning of living events, and these just are questions about a story — about the relation of continually adjusted means to the needs, strivings, and qualities of a particular life. It is no surprise, then, that our answers must be gained in the way we come to understand a story — for example, in the way we make sense of a journey rather than in the way we grasp the physical mechanics of walking.
And then there is water
— the mediator of flow
I have long thought that some day water will be seen as the single most fundamental, “information-rich” physical constituent of life, and that revelations in this regard will outweigh in significance even those concerning the structure of the double helix. Not many biologists today would countenance such a suggestion, and I am not going to mount a serious defense of it here, if only for lack of ability. Time will decide the matter soon enough. But I was particularly pleased to find that the widely read and respected Nature columnist, Philip Ball, once entitled a piece, “Water as a Biomolecule.” In it he wrote:
Water is not simply “life’s solvent,” but rather an active matrix that engages and interacts with biomolecules in complex, subtle and essential ways … Water needs to be regarded as a protean, fuzzily delineated biomolecule in its own right (Ball 2008a; see also Ball 2008b.)
In another paper, Ball (2011) summarized some work bearing on the role of water in biological contexts. The main topic had to do with the relation between water, the binding cavity of an enzyme, and the substrate molecule to which the enzyme binds. It turns out, according to the authors of a study Ball cites, that “the shape of the water in the binding cavity may be as important as the shape of the cavity.” Ball goes on to remark:
Although all this makes for a far more complicated picture of biomolecular binding than the classic geometrical “lock and key” model, it is still predicated on a static or quasi-equilibrium picture. That, too, is incomplete.
Then he cites another paper on enzyme-substrate binding. There it is revealed that, before the binding is complete, water movement near the enzyme is retarded. “Crudely put, it is as if the water ‘thickens’ towards a more glassy form, which in turn calms the fluctuations of the substrate so that it can become locked securely in place. It is not yet clear what causes this solvent slowdown as a precursor to binding; indeed, the whole question of cause and effect is complicated by the close coupling of protein and water motion and will be tricky to disentangle. In any event, molecular recognition here is much more than a case of complementarity between receptor and substrate — it also crucially involves the solvent.”
All this suggests to Ball that “changes in protein and solvent dynamics are not mere epiphenomena, but have a vital role in substrate binding and recognition.”
Structural biologists Mark Gerstein and Michael Levitt (the latter a 2013 Nobel laureate in chemistry) wrote a 1998 article in Scientific American entitled “Simulating Water and the Molecules of Life.” In it they mentioned how early efforts to develop a computer simulation of a DNA molecule failed; the molecule (in the simulation) almost immediately broke up. But when they included water molecules in the simulation, it proved successful. “Subsequent simulations of DNA in water have revealed that water molecules are able to interact with nearly every part of DNA’s double helix, including the base pairs that constitute the genetic code” (Gerstein and Levitt 1998).

Figure 5.3. A representation of the hydration shell of myoglobin, where blue is the myoglobin protein and the small, red-and-white figures stand for water molecules.7
Early attempts to simulate protein molecules rather than DNA produced an analogous difficulty, with the same, water-dependent resolution. Gerstein and Levitt concluded their article with this remark:
When scientists publish models of biological molecules in journals, they usually draw their models in bright colors and place them against a plain, black background. We now know that the background in which these molecules exist — water — is just as important as they are.
That was twenty-five years ago. Today the background remains to be filled in, even if we are now seeing signs of change. Philip Ball (who cites that Gerstein/Levitt remark, and who reproduces two images like the one in Figure 5.3) has more recently noted “an interesting sociological question,” namely, “why certain communities in science decide that particular aspects of a problem are worth devoting a great deal of attention to while others become minority concerns, if not in fact regarded as somewhat suspect and disreputable.” He adds:
Why should we place so much emphasis, for example, on determining crystal structures of proteins and relatively little on a deep understanding of the [water-related] forces … that hold that structure together and that enable it to change and flex so that the molecule can do its job? (Ball 2013)
Certain peculiar historical episodes have contributed to the disreputability of water as a “molecule of life.” (Too many researchers have thought they glimpsed something about water that went beyond current principles of understanding, so that work of this sort came to be seen as mystically tainted or “on the fringe.”) But surely part of the answer to Ball’s question has to do with the longstanding distortion of biology due to the emphasis upon code and mechanism. It is much easier to imagine the step-by-step execution of a computer-like code or the clean insertion of a key into a lock than it is to come to terms with fluid transformations — that is, with what is actually life-like.
The high era of molecular biology that followed upon discovery of “the” structure of the double helix was indeed the Age of Simplicity. We can be thankful that the feverish enchantment of fixed code and crystal is now giving way to an increasing recognition of movement, flow, dynamically flexible interaction, and the continual transfiguration of form — prime narrative elements in the organism’s story.
Organisms Are Activities, Not Things
Many observers have sensed, whether vividly or dimly, that the modern fixation upon things rather than activities — on what has already become rather than the process of becoming — severely distorts our sense of reality. But it is hard for us today to step fully out of this distortion. And nowhere is that distortion more destructive than in the science of life.
Perhaps for that very reason the distortion is also more visible in the science of life. And thanks to new imaging technologies, the visibility is now quite literal. At the cellular level, novel techniques are enabling us to see not only frozen, crystallized structures, but living movement. DNA, RNA, and proteins are being reconceived as “biological soft matter,” subject to continually changing form so that molecular performances become more like improvised dances than automatic lock-and-key mechanical interactions. “Disordered” or “unstructured” sequences in proteins are now seen as decisive for coordinated activities throughout the cell, from gene regulation to signaling across membranes.
Still more dramatically, molecular biologists have in recent years become almost transfixed by the novel importance of phase transitions — for example, the forming and dissolving of distinctive, membraneless droplets within the fluid cell, whereby specialized and localized functional capacities are maintained despite the rapid passage of molecules in and out of the droplets.
And perhaps most important of all is the nascent recognition — which still hasn’t taken widespread hold in biology — that the amazing functional plasticity of water may be key to just about everything that goes on in a cell.
All this points us to the question of coherence: how are the virtually infinite “degrees of freedom,” so evident in the free flows of the cell, disciplined and subordinated to the larger purposes of the cell, whether they be gene expression or intercellular communication or metabolism or cell division. In the next chapter (“Context: Dare We Call It Holism?”) and in Chapter 8 (“The Mystery of an Unexpected Coherence”) we will try to get some clearer views of this larger, meaningful picture.
Notes
1. The twentieth-century American philosopher, Susanne Langer, clearly grasped the essence of the matter in her own discussion of the heart’s development and functioning. The heart, she said,
begins to form early in embryonic life, apparently serving no purpose until the incipient vascular system is ready to act with it. In the earliest phases, however, a characteristic function of periodic contraction, the so-called “pulse,” appears in many evolving tissues, some of which will cease to exhibit it later, while others will join the cardiac development, so their rhythms will become entrained by larger ones and finally by the [entire] circulatory pulse.
This preliminary beating, which comes early in the heart’s formation, “illustrates a basic characteristic of organic function, namely, that its integated activities are often detectable before their special mechanisms have even begun to appear.” This is a powerful reminder that, in an organism’s development, the part “descends from,” or is differentiated within, its larger context, which is ultimately the whole organism. Speaking further of the heart’s development, Langer wrote:
Nothing could demonstrate more aptly the primacy of acts in biological existence, and their gradual concentration in those regions of an organism where they can expand, dominate and integrate most fully. This order of development, from differentiating function to specialized location (tissue determination) and finally specialized form (cell determination), has been noted many times by embryologists. [American zoologist] Charles Manning Child remarked, fifty years ago, that “differences in reaction or in capacity to react very commonly exist in different parts even before visible differentiation occurs, or in cases where it never occurs.”
Langer reinforces these remarks by citing the embryologist and author of Form and Causality in Early Development, Albert M. Dalcq, to the effect that, to begin with, the unity of the nervous system “is not so much spatial as functional … The nervous system does not really originate from a unique and continuous layer of cells.” And the American developmental biologist, Clifford Grobstein, whose life spanned much of the twentieth century, concluded from his experimental studies of development in young embryos that “when nervous tissue ‘self-differentiates’ … the cells themselves have not yet acquired fixity of type as nerve cells. … some stabilization at the tissue level seems to precede stabilization at the cell level” (Langer 1967, pp. 200, 401-2).
For a more recent discussion of the heart, see the impressive evidences and analysis in Branko Furst’s technical treatise on The Heart and Circulation: An Integrative Model (Furst 2020).
2. Figure 5.1 credit: Courtesy of Margot Quinlan (copyright).
3. A terminological issue: Turoverov and colleagues speak more specifically of “highly dynamic biological soft matter positioned at the edge of chaos.” The abstract and perhaps rather tiresome notion of “the edge of chaos” is better captured in this context by a picture of life-like processes — powerfully organized, but in a dynamic manner that continually adapts to circumstances from a purposive, and therefore not physically predictable, center of agency. The predictability, such as it is, lies in the reasonable expectation of coherence in the interweaving meanings we observe. (See Chapters 2 and 8.
4. Biologists often speak of communication in terms of signals and signaling, where signal can hardly be distinguished in any absolute way from cause. However, “signals” tend to be spoken of where there are repeated, more or less stereotypical sequences (“pathways”) of molecular interaction between different cells, leading to more or less consistent consequences. This happens, for example, when a gland secretes a hormone (“signal”) that subsequently has effects in other parts of the body.
Wikipedia offered this definition of “cell signaling” in August, 2019: “Cell signaling is part of any communication process that governs basic activities of cells and coordinates multiple-cell actions. The ability of cells to perceive and correctly respond to their microenvironment is the basis of development, tissue repair, and immunity, as well as normal tissue homeostasis.” This easy acknowledgment of “communication,” “coordination,” “governance,” “perception,” and “correct response” — all within a science that, on the surface, refuses the normal and unavoidably immaterial meaning of these terms — illustrates the biologist’s blindsight described in Chapter 1.
5. A sol-gel transition occurs when a solution (in which one substance is dissolved in another) passes into a gel state. The latter consists of a solid molecular lattice that is expanded throughout its volume by a fluid — water, in the case of a hydrogel. The fluid may constitute over 99% of the volume of the gel, yet the solid lattice prevents the gel from flowing like a liquid.
6. Here is one of innumerable examples of the role of phase separation in physiological processes: “Cells under stress must adjust their physiology, metabolism, and architecture to adapt to the new conditions. Most importantly, they must down-regulate general gene expression, but at the same time induce synthesis of stress-protective factors, such as molecular chaperones … [We] propose that the solubility of important translation factors is specifically affected by changes in physical–chemical parameters such [as] temperature or pH and modulated by intrinsically disordered prion-like domains. These stress-triggered changes in protein solubility induce phase separation into aggregates that regulate the activity of the translation factors and promote cellular fitness” (Franzmann and Alberti 2019).
7. Figure 5.3 credit: From Frauenfelder et al. 2009.
Sources
Ball, Philip (2008a). Water as a Biomolecule, ChemPhysChem vol. 9, pp. 2677-85. doi:10.1002/cphc.200800515
Ball, Philip (2008b). Water as an Active Constituent in Cell Biology, Chemical Reviews vol. 108, no. 1, pp. 74-108. doi:10.1021/cr068037a
Ball, Philip (2011). More Than a Bystander, Nature vol. 478 (October 27), pp. 467-68. doi:10.1038/478467a
Ball, Philip (2013). Concluding Remarks: Cum Grano Salis, Faraday Discussions vol. 160, pp. 405–14. doi:10.1039/c2fd20126g
Dawkins, Richard (2006). The Blind Watchmaker, third edition. New York: W. W. Norton. First edition published in 1986.
Dolgin, Elie (2022). The Shape-Shifting Blobs That Rule Biology, Nature vol. 611 (November 3), pp. 24-27. doi:10.1038/d41586-022-03477-y
Dunker, A. Keith, Christopher J. Oldfield, Jingwei Meng et al. (2008). The Unfoldomics Decade: An Update on Intrinsically Disordered Proteins, BMC Genomics vol. 9 (Suppl 2):S1. doi:10.1186/1471-2164-9-S2-S1
Emerson, R. W. (1908). The Conduct of Life: Nature and Other Essays. New York: E. P. Dutton. Available at https://archive.org/details/conductoflifenat00emeriala
Franzmann, Titus M. and Simon Alberti (2019). Protein Phase Separation as a Stress Survival Strategy, Cold Spring Harbor Perspectives in Biology vol. 11, no. 6 (June). doi:10.1101/cshperspect.a034058
Frauenfelder, Hans, Guo Chen, Joel Berendzen et al. (2009). A Unified Model of Protein Dynamics, PNAS vol. 106, no. 13 (March 31), pp. 5129-34. doi:10.1073/pnas.0900336106
Furst, Branko (2020). The Heart and Circulation: An Integrative Model, 2nd edition. New York: Springer.
Ganser, Laura, Megan L. Kelly, Daniel Herschlag and Hashim M. Al-Hashimi (2019). The Roles of Structural Dynamics in the Cellular Functions of RNAs, Nature Reviews Molecular Cell Biology (Aug). doi:10.1038/s41580-019-0136-0
Gerstein, Mark and Michael Levitt (1998). Simulating Water and the Molecules of Life, Scientific American vol. 279, no. 5 (November), pp. 100-5. doi:10.1038/scientificamerican1198-100
Gerum, R. C., B. Fabry, C. Metzner et al. (2013). The Origin of Traveling Waves in an Emperor Penguin Huddle, New Journal of Physics vol. 15 (December). https://iopscience.iop.org/article/10.1088/1367-2630/15/12/125022
Grant, Barry J., Alemayehu A. Gorfe and J. Andrew McCammon (2010). Large Conformational Changes in Proteins: Signaling and Other Functions, Current Opinion in Structural Biology vol. 20, pp. 142-47. doi:10.1016/j.sbi.2009.12.004
Gsponer, Jörg and M. Madan Babu (2009). The Rules of Disorder Or Why Disorder Rules, Progress in Biophysics and Molecular Biology 99, no. 2-3 (February-May), pp. 94-103. doi:10.1016/j.pbiomolbio.2009.03.001
Halfmann, Randal (2016). A Glass Menagerie of Low Complexity Sequences, Current Opinion in Structural Biology vol. 38, pp. 18-25. doi:10.1016/j.sbi.2016.05.002
Harold, Franklin M. (2001). The Way of the Cell: Molecules, Organisms and the Order of Life. Oxford: Oxford University Press.
Hilser, Vincent J. (2013). Structured Biology: Signalling from Disordered Proteins, Nature vol. 498 (June 20), pp. 308-10. doi:10.1038/498308a
Holdrege, Craig, editor (2002). The Dynamic Heart and Circulation. The chapters originally written in German were translated by Katherine Creeger. Fair Oaks CA: AWSNA.
Hubstenberger, Arnaud, Scott L. Noble, Cristiana Cameron and Thomas C. Evans (2013). Translation Repressors, an RNA Helicase, and Developmental Cues Control RNP Phase Transitions during Early Development, Developmental Cell vol. 27 (October 28), pp. 161-73. doi:10.1016/j.devcel.2013.09.024
Hyman, Anthony A. and Clifford P. Brangwynne (2011). Beyond Stereospecificity: Liquids and Mesoscale Organization of Cytoplasm, Developmental Cell vol. 21 (July 19), pp. 14-16. doi:10.1016/j.devcel.2011.06.013
Landecker, Hannah (2012). The Life of Movement: From Microcinematography to Live-Cell Imaging, Journal of Visual Culture vol. 11, pp. 378-99. doi:10.1177/1470412912455622
Pearson, Helen (2003). Beyond the Double Helix, Nature vol. 421 (January 23), pp. 310-12. 10.1038/421310a
Rothman, Stephen (2002). Lessons from the Living Cell: The Limits of Reductionism. New York: McGraw Hill.
Schad, Wolfgang (2002). A Dynamic Morphology of the Cardiovascular System, in Holdrege (2002), pp. 77-97.
Shin, Yongdae and Clifford P. Brangwynne (2017). Liquid Phase Condensation in Cell Physiology and Disease, Science vol. 357, no. 6357 (September 22). doi:10.1126/science.aaf4382
Szewczak, Lara (2019). Just Solid or Liquid Enough, Cell vol. 178, no. 4 (August 8), pp. 763-64. doi:10.1016/j.cell.2019.07.027
Talbott, Stephen L. (2024). How Do Biomolecules “Know” What To Do? https://bwo.life/bk/ps/biomolecules.htm
Tompa, Peter (2013). Hydrogel Formation by Multivalent IDPs: A Reincarnation of the Microtrabecular Lattice?, Intrinsically Disordered Proteins 1:e24068 (January–March). doi:10.4161/idp.24068
Turoverov, Konstantin K., Irina M. Kuznetsova, Alexander V. Fonin et al. (2019). Stochasticity of Biological Soft Matter: Emerging Concepts in Intrinsically Disordered Proteins and Biological Phase Separation, Trends in Biochemical Sciences vol. 44, no. 8 (August 1), pp. 716-28. doi:10.1016/j.tibs.2019.03.005
Zhou, Yaoqi, Dennis Vitkup and Martin Karplus (1999). Native Proteins Are Surface-Molten Solids: Application of the Lindemann Criterion for the Solid versus Liquid State, Journal of Molecular Biology vol. 285, pp. 1371-1375. doi:10.1006/jmbi.1998.2374
Zhu, Lian and Clifford P. Brangwynne (2015). Nuclear Bodies: The Emerging Biophysics of Nucleoplasmic Phases, Current Opinion in Cell Biology vol. 34, pp. 23-30. doi:10.1016/j.ceb.2015.04.003
Steve Talbott :: Our Bodies Are Formed Streams
