What Brings Our Genome Alive?
Throughout most of the twentieth century, genes were viewed as the “agents” responsible for an organism’s development, activity, and evolution. Their agency was said to lie in their ability to “regulate,” “organize,” “coordinate,” and “control” physiological processes, and their changes (“mutations”) were the material of evolution. DNA, the bearer of these genes, became the “Book of Life” — the essential maker of organisms and driver of evolution. And this view remains stubbornly entrenched today, despite many changes in our understanding. In 2019 a leading behavioral geneticist could still write a book titled, Blueprint: How DNA Makes Us Who We Are.
Nevertheless, the idea that genes are the decisive “first causes” of life — and, more generally, that molecules at the “bottom” ultimately explain everything that happens at larger scales — has come in for a great deal of criticism in recent years. This criticism, as we will see, is fully justified. But the issues can be subtle, as is suggested by an apparent paradox. Philosopher of biology Lenny Moss, who wrote the valuable book, What Genes Can’t Do, has remarked:
Where molecular biology once taught us that life is more about the interplay of molecules than we might have previously imagined, molecular biology is now beginning to reveal the extent to which macromolecules [such as DNA], with their surprisingly flexible and adaptive complex behavior, turn out to be more life-like than we had previously imagined (Moss 2012).
In a similar vein, I myself have written:
Having plunged headlong toward the micro and molecular in their drive to reduce the living to the inanimate, biologists now find unapologetic life staring back at them from every chromatogram, every electron micrograph, every gene expression profile. Things do not become simpler, less organic, less animate. The explanatory task at the bottom is essentially the same as what we faced higher up (Talbott 2010).
But if this is really true, what are we to make of Harvard geneticist Richard Lewontin’s declaration, itself hardly disputable, that
DNA is a dead molecule, among the most nonreactive, chemically inert molecules in the living world. That is why it can be recovered in good enough shape to determine its sequence from mummies, from mastodons frozen tens of thousands of years ago, and even, under the right circumstances, from twenty-million-year-old fossil plants.
Many astute observers have echoed Lewontin’s remarks, and I have never seen anyone question them, including those who remain enamored of the “Book of Life.” So which is it? When we peer at DNA, do we see a dead molecule or a living dynamic? Lewontin himself, in that same passage, pointed toward the answer (we will try to forgive his ill-fitting use of the word “machinery”):
DNA has no power to reproduce itself. Rather it is produced out of elementary materials by a complex cellular machinery of proteins. While it is often said that DNA produces proteins, in fact proteins (enzymes) produce DNA … Not only is DNA incapable of making copies of itself, aided or unaided, but it is incapable of “making” anything else (Lewontin 1992).
In other words, the proper functioning of DNA is an achievement of its entire cellular context. If we conceive the molecule in the usual way as a bit of mindless, inherently inert matter, then, according to our own conceptions, we see only dead stuff. But if we conceive the molecule as a system of forces and energies capable of participating in, and being caught up in, the creative life of the whole cell and organism, then we can hardly help recognizing — and perhaps even reverencing — the living performance unfolding before our eyes.
Saying this is one thing; making it both meaningful and profound is quite another — and that is one task of the present book.
The genome as you have
probably not heard about it
If you arranged the DNA in a human cell linearly, it would extend for nearly two meters. How do you pack all that DNA into a cell nucleus just five or ten millionths of a meter in diameter? According to the usual comparison it’s as if you had to cram twenty-four miles (thirty-nine kilometers) of extremely thin thread into a tennis ball. Moreover, this thread is divided into forty-six pieces (individual chromosomes) averaging, in our tennis-ball analogy, over half a mile long. Can it be at all possible not only to fit those chromosomes in the nucleus, but also to keep them from becoming hopelessly entangled?
Box 3.1
Some Standard Terminology
The usual formula has it that DNA makes RNA and RNA makes protein. The DNA double helix forms a kind of spiraling ladder, with pairs of nucleotide bases constituting the rungs of the ladder: a nucleotide base attached to one siderail of the ladder bonds with a base attached to the other siderail. These two bases, commonly referred to as “base pairs” (“letters” of the DNA “text”), are normally complementary, so that, of the four different bases (abbreviated as A, T, C, and G), an A pairs only with a T (and vice versa), just as C and G are paired. Each siderail, with all its attached nucleotide bases, is considered a single strand of the double helix. Because the chemical subunits making up the siderails are asymmetrical and oriented oppositely on the two strands, the strands can be said to “point” in opposite directions.
The enzyme that transcribes DNA into RNA (thereby expressing a gene) must move along the length of the gene in the proper direction, separating the two strands and using one of them, with its sequence of nucleotide bases, as a template for synthesizing a single-stranded RNA transcript — a transcript that complements the template DNA strand in much the same way that one DNA strand complements the other. It is by virtue of this complementation that the “code” for a protein is said to be passed from DNA to RNA. Once synthesized, the RNA may pass through the nuclear envelope to the cell’s cytoplasm, where it may be translated into protein.
It all makes for a neat, if (as told here) greatly simplified, story. For a fuller exploration of technical terms, see the glossary at https://bwo.life/mqual/glossary.htm.
Obviously it must be possible, however difficult to conceive. The first thing to realize is that chromosomes do not consist of naked DNA. Their actual substance, an intricately woven and ever-changing structure of DNA, RNA, protein, and other molecules, is referred to as chromatin. (See Box 3.1 for some basic terminology.) Histone proteins, several of which can bind together in the form of a complex histone core particle, are the single most prominent, non-DNA constituents of this chromatin. Every cell contains numerous such core particles — there are some 30 million in a typical human cell — and the DNA double helix, after wrapping a couple of times around one of them, typically extends for a short stretch and then wraps around another one. The core particle with its DNA wrapping is referred to as a nucleosome (about which you can read much more in Chapter 14, “How Our Genes Come to Expression”), and between 75 and 90 percent of our DNA is said to be wrapped up in nucleosomes. This is one way the cell packs its DNA into a surprisingly small volume.
But how is all this material organized so as to serve the infinitely complex requirements of a flatworm, bumblebee, shark, or human? Biologists have spent a good number of years trying to visualize the organization of chromosomes in the cell nucleus, and, unsurprisingly, the picture tends to differ depending on the scale at which you look.1 Most broadly, the genome appears to be partitioned into two compartments, the “A” compartment, with more “open” (less densely packed) chromatin and more active genes, and the “B” compartment, with more “closed” chromatin and less active genes. Some researchers have pointed to the existence of several subcompartments distinguished by the presence of distinctive features (“marks”) on the proteins associated with the DNA.
At a somewhat smaller, megabase scale, there are so-called “topologically associated domains,” within which the interactions among loci are more frequent than across such domains. Also at this scale, it is now thought that some chromosome regions form “fractal globules” that are more or less free of knots (Figure 3.1).
And, at a still smaller scale (roughly 200,000 bases), there are loop domains generally associated with active genes (Figure 3.2). For a smaller scale yet — one that is intensely relevant to gene regulation and expression — see the discussion of nucleosomes in Chapter 14.
How all this fits together is, of course, less than fully clear. And things only become more complex when you consider that loci on separate chromosomes often come into complex and intimate relation with each other, in part because of the need to coordinate the expression of genes on different chromosomes. And there are also the chromatin proteins, the modifications of those proteins, and the vast number of associated molecules in the nucleus that influence how genes will be expressed.


Figure 3.1. A schematic representation of a proposal for how parts of a human chromosome can be organized into an unknotted “fractal globule” in the cell nucleus. The linear chromosome segment at the top of the figure shows, in a miniaturized way, what the unfolded globule might look like.2
The image shown in Figure 3.1 is a geometric idealization. It is designed to show certain principles of the folding of chromosomes at the megabase scale, and is not meant to suggest that any part of any chromosome is organized into a neat sphere.
In reality, the cell nucleus presents us with an almost infinitely complex and dynamic configuration of functionally related regions all the way down to the smallest scale. Different parts of the same chromosome might lie in the “A” and “B” compartments and can move between them. Similarly, loops can form and disappear. And nucleosomes, we will see in Chapter 14, seem almost continually in movement, which is central to gene expression.
There is also continual engagement between the genome and other contents and activities of the nucleus. For example, substantial portions of the “B” compartment reside near, and interact with, the outer envelope of the nucleus, whereas much of the “A” compartment lies more in the interior. During the processes of DNA replication and cell division (mitosis), the entire arrangement, for all its seemingly convoluted complexity, radically transforms into a series of different configurations. (See, for example, Figure 3.3.)
The picture is always dynamic. But it’s not so much that chromosomes move as that they are brought into movement. Particular genes — which is to say, particular parts of chromosomes — can be shifted from one place to another, and the associations thereby formed with other chromosomal regions, whether on the same or different chromosomes, can be decisive for the regulation of gene expression. We can easily wonder how the overall choreography of the cell nucleus and whole cell can be perfectly “calculated” for the management of the 20,000 genes and millions of significant loci in the genome. And the intricately dynamic relationships between different chromosomes give us a glimpse of how misleading an image like that of Figure 3.1 can be, with its geometrically compact, isolated, and static character.
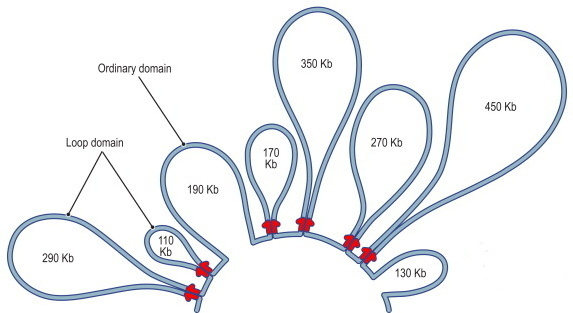
Figure 3.2. A schematic representation of DNA loops. “Kb” stands for “kilobases,” or thousands of “letters” of the “genetic code” — in this case, the number of letters strung out along the length of a chromosome loop. For example, the loop at far left is 290,000 letters long.3 (On terminology, see Box 3.1.)
In Figure 3.2 the paired red marks at the point where a loop converges on itself indicate the presence of two copies of a particular protein, one of a number of molecules that play a role in loop formation. (How do they “know” where to place themselves?) Of the two widely separated loci thus brought together, one may be near a gene while the other is near DNA regulatory sequences necessary for that gene to be expressed. Their coming together (or not) is therefore part of how particular genes come to be expressed (or not). And, likewise, the reconfiguration of such loops may be critical for the altered expression of genes as the cellular and organismal context changes.
Note that the two loci where the protein binds a particular loop can be separated on the linear chromosome by hundreds of thousands of genetic “letters.” (For comparison, while genes vary greatly in size, they average about 30,000 “letters” in length. And human chromosomes range from about 47 to 247 million “letters.”)
So we have seen that there are different ways for genes to be brought into “community,” all of which becomes extremely complex, as it surely must, given the diverse uses to which radically distinct cell types must put some of their genes. Investigations into the organization of chromosomes for different functions and at different scales can probably be said to be at an early stage, and the picture will doubtless become still more complex as research proceeds. At present there seem to be no absolute rules of interaction, and the question of clear-cut cause and effect always seems to be in doubt (Chapter 9, “A Mess of Causes”). For example, highly expressed genes are strongly associated with chromosome loops, but they do not absolutely have to be.
Most of the foregoing description has been more or less static. We have so far hardly done more than hint at the true dynamism that enlivens our genetic heritage, but we have perhaps already glimpsed that gesture in three-dimensional space is crucial. And the general picture of the genome’s dynamic spatial organization has seemed to galvanize molecular biologists. John Rinn, director of the Rinn Lab at Harvard, has said of the nuclear space and its chromosomal drama, “It’s genomic origami … It’s the shape that you fold [the genome] into that matters” (quoted in Zimmer 2015).
According to a paper from another group of researchers, “A loop that turns a gene on in one cell type might disappear in another. A domain may move from subcompartment to subcompartment as its flavor changes. No two cell types [have their chromosomes] folded alike. Folding drives function.”4 Or, as we might put it, gesturing gives expressive shape to the cell’s life. Suhas Rao, the paper’s lead author and a researcher at Baylor College of Medicine’s Center for Genome Architecture, remarked:
A loop is the fundamental fold in the cell’s toolbox. We found that the formation and dissolution of DNA loops inside the nucleus enables different cells to create an almost endless array of distinct three-dimensional folds and, in so doing, accomplish an extraordinary variety of functions (quoted in Physorg 2014).
Every overall configuration of chromosomes in the nucleus (involving many factors we have not yet considered) represents a unique combination of expressed and repressed genes among our total complement of 20,000 or so genes.5 Moreover, new features of chromosome spatial and dynamic organization continue to be elucidated on a regular basis, and there appears to be no limit to the variety and scale of these features.
Think about all this dynamic form and movement for a while, and you may find yourself asking, along with me: What possible “mechanism” could ensure the coherence of all this movement and gesturing in relation to all the requirements of the trillions of cells in your or my body, or the tissues and organs into which those cells are organized, as we go about our endlessly varying activities under endlessly varying conditions?
Of dynamism and mystery
in the cell nucleus
The chromosome, remarked Christophe Lavelle of France’s Curie Institute, “is a plastic polymorphic dynamic elastic resilient flexible nucleoprotein complex.”6 There are many activities in which it is caught up, revealing significant form and organization. In order to visualize just one of these activities, consider a long, double-stranded rope whose two strands coil around each other, much like the two strands of a DNA molecule. If you twist a segment of this rope in a manner opposite to its natural spiraling, you will find that the strands tend to separate (that is, loosen, or become less tightly wound). And if you continue to twist, then the rope as a whole will begin to coil upon itself (called “negative supercoiling”). Similarly, if you twist in the same direction as the rope’s natural twist, you will tighten the winding of the strands, and if you continue twisting, the rope will again coil upon itself (“positive supercoiling”).
The DNA double helix can likewise be loosened by twisting, along with formation of coils, and it can also be tightened and coiled. In fact, it happens that both effects result wherever the enzymes transcribing DNA into RNA are at work. And this twisting in one direction or another in turn either encourages or discourages the expression of nearby genes.7
In other words, in addition to the chromosome domains discussed above, there are transient domains established by the twisting (torsional) forces that are communicated more or less freely (and not only by transcribing enzymes) along bounded segments of the chromosome. The loci within such a region share a common torsion, and this can attract a common set of regulatory proteins that read the changes as “suggestions” about activating or repressing nearby genes (Lavelle 2009; Kouzine et al. 2008). The torsion also tends to correlate with the level of compaction of the chromatin fiber, which in turn correlates with many other aspects of gene regulation.
Picture the situation concretely. Every bodily activity or condition presents its own requirements for gene expression. Whether you are running or sleeping, starving or feasting, rousing yourself to action or calming down, suffering a flesh wound or recovering from pneumonia — in all cases the body and many of its different cells have specific, almost incomprehensibly complex and changing requirements for differential expression of thousands of genes. And one thing (among countless others) bearing on this differential expression in all its fine detail is the coiling and uncoiling of chromosomes.
With so much concerted movement going on (including the looping we heard about earlier) how does the cell keep all those “twenty four miles of string in the tennis ball” from getting impossibly tangled? We do at least know some of the players addressing the problem. For example, there are complex protein enzymes called topoisomerases, which the cell employs to help manage the spatial organization of chromosomes. Demonstrating a spatial insight and dexterity that might amaze those of us who, even with the benefit of full consciousness, have struggled to sort out tangled masses of thread, these enzymes manage to make just the right local cuts to the strands in order to relieve strain, allow necessary movement of individual genes or regions of the chromosome, and prevent a hopeless mass of knots.
Some topoisomerases cut just one strand of the double helix, allow it to wind or unwind around the other strand, and then reconnect the severed ends. This alters the coiling of the DNA. Other topoisomerases can undo knots by cutting both strands, passing a loop of the chromosome through the gap thus created, and then sealing the gap again.
Imagine trying this with miles of string wrapped around millions of minuscule beads compacted into a few cubic inches of space (tennis ball), with the string all the while looping and squirming like a nest of snakes in order to bring all the right loci together so as to achieve the tasks of the moment. (And how are these tasks “known”?) I don’t think anyone would claim to have the faintest idea how this is actually managed in a meaningful, overall, contextual sense, although great and fruitful efforts have been made to analyze the local forces and “mechanisms” at play in isolated interactions.
Does the lawfulness of
molecular interactions
explain global coherence?
We have scarcely begun to look at the dynamic aspects of the cell nucleus. Not only are chromosomes made to fold, loop, coil, and twist rather like a nest of snakes, but they engage in decisive and changing electrical interactions; they relocate from here to there within the nucleus, partly in order to associate with dynamically assembled collections of molecules important for regulating gene expression; and they are influenced by pushes and pulls from the fibers of the extra-nuclear cytoskeleton (Chapter 4, “The Sensitive, Dynamic Cell”).
Or again, DNA is said to “breathe” in rhythmical movements as it tightens and relaxes its embrace of the histone core particles mentioned earlier. And again, it breathes in a different way and in a different sort of rhythm as lengths of the two strands of the double helix alternately separate and reunite. And yet again, there are many profoundly significant structural novelties to which DNA lends itself, beyond the conventional form of the double helix. All this and much more is the cell’s way of evoking the genetic performance that it needs — a performance that expresses the cell’s own life and that of the organism as a whole.8
And so, when researchers refer to the “choreography” of the cell nucleus and the “dance” of chromosomes, as they sometimes do, their language is closer to being literal than many have imagined. If the organism is to survive, chromosomal movements must be well-shaped responses to sensitively discerned needs — all in harmony with innumerable dance partners, and all resulting in every gene being expressed or not according to the meaning of the larger drama. We can hardly help asking: If such a qualitative choreography is how the organism lives and performs at the molecular level, what does this mean for the nature of molecular biological explanation — especially when we are acknowledging an organism’s qualitative needs, interests, and purposes?
Yes, the use of terms such as “dance” and “choreography” in molecular biology is rather distinctive. Some might call it eccentric. But this particular eccentricity has for some time now been creeping into the conventional technical literature. We have already heard of “genomic origami.” And we have also been told: “The statement, ‘genomes exist in space and time in the cell nucleus’ is a trivial one, but one that has long been ignored in our studies of gene function” — this according to two leaders of the current work: Job Dekker, head of a bioinformatics lab studying the spatial organization of genomes at the University of Massachusetts Medical School, and Tom Misteli, a research director at the National Cancer Institute. Recent investigations, they say, have taught us that “gene expression is not merely controlled by the information contained in the DNA sequence,” but also by the “higher-order organization of chromosomes” and “long-range interactions in the context of nuclear architecture” (Dekker and Misteli 2015).
This last remark may startle some readers into the sudden realization that in all the foregoing there has been scant mention of the famed DNA sequence — the supposedly precise logical content of the “coded genetic program” that “makes us who we are.” Why is that?
It looks very much as if the chromosome, along with everything else in the cell, is itself a manifestation of life, not a logic or mechanism explaining life. This performance cannot be captured with an abstract code. Gene regulation is defined less by static elements of logic than by the quality and force of the cell’s gesturing as it brings its genome into movement. The chromosome becomes an expression of a larger context of living activity. As Nature columnist Philip Ball has put it, the clean logic of the DNA code, as it has been commonly formulated, “is so elegant that it risks blinding us to the awesome sophistication of the total process” (Ball 2003).
The fixation upon an abstract, neatly identifiable informational sequence has served well the aim of biologists to find precise, unambiguous, logically clean, and satisfyingly deterministic causal explanations. Nevertheless, what’s been happening in rapidly intensifying fashion over the past couple of decades, has been a forced retreat from explanations of this sort. To cite a few key words and phrases from the contemporary literature: everything turns out to be mind-numbingly complex, which means, in part, that context makes all the difference. We are forced to try to understand how regulatory networks, intricate feedback loops, and the frequent difficulty of distinguishing causes from effects bear upon our biological understanding. Ultimately, we seem to be driven toward systems biology, an easily degraded term that many seem to prefer over the embarrassment (and richer meaning) of holistic biology.
What is not generally realized, however, is that this retreat from simplistic “causal mechanisms” suggests a movement toward a kind of explanation biologists have not yet come to terms with. It is, after all, one thing to explain, say, how a topoisomerase enzyme “mechanistically” passes one double-stranded section of DNA through another, and quite a different thing to ask how this activity — which could be carried out in countless different patterns — is made to harmonize with everything else going on at the molecular level in order to produce an overall, directed, coherent outcome for the cell as a whole. How might we make sense of the vast coordination of trillions of molecular events in the interest of a larger picture that is subject to continual change, as when a cell initiates the transition leading toward cell division (which changes the meaning of everything going on)?
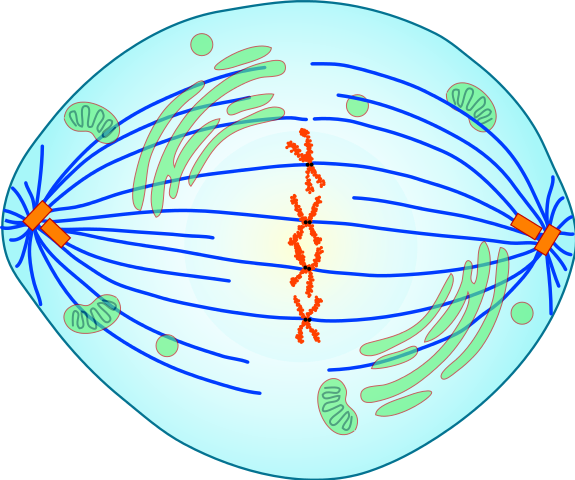
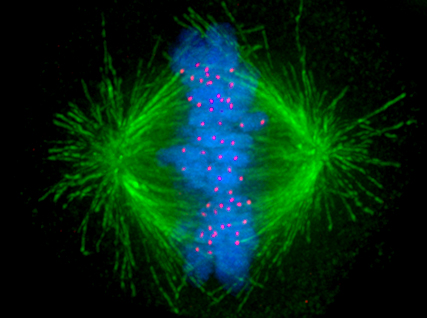
Figure 3.3. (top:) A schematic representation of a mitotic spindle in a cell with just four duplicated chromosomes. (bottom:) An artificially colored image of the mitotic spindle in a human cell, showing microtubules in green, chromosomes in blue, and kinetochores in red. A kinetochore is a protein structure that temporarily holds a chromosome and its duplicate together while also providing an anchor for a “thread” of the mitotic spindle. In the following phase of mitosis, each chromosome and its duplicate will be pulled apart, destined for different daughter nuclei.9
The globular and peculiarly organized aggregation of chromosomes we saw in Figure 3.1 is a long way, for example, from the the chromosomal organization during DNA replication, and likewise from the striking configurations we observe with the mitotic spindle during cell mitosis (Figure 3.3). What is a topoisomerase to do when it is in contact with a particular locus of a DNA molecule — a particular locale among the intricately folded, 6.4 billion nucleotide bases (“letters”) of a human cell? How does it connect with the larger drama, so as to play its local role properly? Or is it rather that the larger drama connects immaterially with the individual topoisomerase?10
James Wang, the Harvard University molecular biologist who discovered the first topoisomerase, seems to have had some awareness of the problem. Writing about the striking capability of a topisomerase to untie a DNA knot by cutting through the double helix and later putting it back together again — all without disturbing the critical continuity of the original chemical structure — he expresses his wonder:
When we think a bit more about it, such a feat is absolutely amazing. An enzyme molecule, like a very nearsighted person, can sense only a small region of the much larger DNA to which it is bound, surely not an entire DNA [molecule]. How can the enzyme manage to make the correct moves, such as to untie a knot rather than make the knot even more tangled? How could a nearsighted enzyme sense whether a particular move is desirable or undesirable for the final outcome? (Wang 2009)
Despite his language, Wang presumably knows that a molecule does not sense anything at all. And he surely also knows that the topoisomerases always have an adequate physical basis for doing what they do in the place where they are. And yet this physically lawful activity (which is what Wang concerns himself with) does not yet get us to an understanding of how the enzymes act in support of radically different purposes as a cell proceeds through DNA replication, for example, or gene transcription, or the distinctive phases of cell division.
Wang’s reference to whatever is “desirable or undesirable for the final outcome” is what we must ultimately reckon with. That is, the context to which the topoisomerase molecule must conform is, in the end, the activity of the whole organism, with its requirements for specific gene expression in every part of the body. Put simply, the molecule must meaningfully participate in everything — organism and environment — without fixed limit.
This points to the need for a kind of explanation biologists in general seem unwilling to acknowledge. For it would, indeed, upset the entire world of conventional biological thought, based as that thought is on local, analyzable, physical cause and effect. “Desirable for the entire context” and, similarly, “undesirable” are not physical categories.11 Yet here is a perfectly competent physical scientist driven to use such phrases. We should pay attention.
Yes, we have every reason to believe that whatever happens, happens lawfully. But this still leaves us with the question, “How does our understanding of the overall coherence of cellular and organismal processes relate to the lawfulness we unfailingly observe whenever we isolate particular interactions and analyze them in physical and chemical terms?” (Talbott 2024) That lawfulness continues the same throughout all cellular activity of the most diverse sorts, and it does not seem to have any obvious provisions for explaining the unique, ever-varying principles of coordination and coherence governing biological entities ranging from cells to organs to whole organisms to different species within their environments.
Yes, the Cell’s Genomic Performance Is Complex!
Where Francis Crick and James Watson (known as the discoverers of the structure of DNA) were looking for a single, univocal code, we now know that a thousand different things are going on. Not only is all the regulatory activity and the resulting, three-dimensional “dance” of our genome exceedingly complex, it also shows us clearly that we are really looking at a whole-cell and whole-organism performance. The genome can do nothing of itself — not even twist itself into coils or “go loopy” — and, in achieving such things, the cell comes at the genome from every possible direction and temporally varies its approach in tune with ever-changing conditions. We will learn more about this complexity in further chapters, especially Chapters 7 (“Epigenetics: A Brief Introduction”) and 14 (“How Our Genes Come to Expression”). The question how everything is coordinated in a useful, need-fulfilling, and meaningful way seems continually to encourage biologists to transcend conventional scientific descriptive language, as when they refer to the “three-dimensional dance of chromosomes.”
You will have noticed in these first chapters that we seem to be raising a lot of questions! You can count on one thing — the question-raising will never come to an end. This is, in the first place, what all good science should do — raise decisive questions with ever greater clarity. But we can also nourish a hope that is not common in today’s science: namely, that by continuing to describe the life of organisms in a revelatory way — acknowledging the narrative and holistic character of beings whose lives manifest from the immaterial “inner” toward the material “outer” — we will find the description itself coming more and more to constitute exactly the sort of biological understanding and explanation we can best look for. We will explicitly address this sort of understanding, and how it connects to our ideas of causality, in Chapter 12 (“Is a Qualitative Biology Possible?”).
We will also confront — especially in Chapters 13 (“All Science Must Be Rooted in Experience”) and 24 (“Is the Inanimate World an Interior Reality?”) — how our questions relate to the problem of the thought-infused character of the material world generally. And while just about the whole book raises a question about the relation between isolated and specific living processes, on one hand, and their larger context, on the other, we will try to make this question more pointed in Chapter 6 (“Context: Dare We Call It Holism?”) and Chapter 8 (“The Mystery of an Unexpected Coherence”).
Notes
1. Two important efforts to map the spatial arrangement of chromosomes were published in 2009 and 2014: Lieberman-Aiden et al. 2009 and Rao et al. 2014.
2. Figure 3.1 credit: Copyright Miriam Huntley, Rob Scharein, and Erez Lieberman-Aiden, from Lieberman-Aiden et al. 2009. Linear chromosome at top of figure: Ed Yong (https://commons.wikimedia.org/wiki/File:Fractal_globule.jpg), CC BY-SA 3.0.
3. Figure 3.2 credit: from Rao et al. 2014.
4. Rao et al. 2014. The quote comes from the authors’ video abstract of their paper in Cell.
5. Toward the end of the Human Genome Project in 2000, according to a report in Nature, “geneticists were running a sweepstake on how many genes humans have, and wagers ranged from tens of thousands to hundreds of thousands. Almost two decades later, scientists armed with real data still can’t agree on the number.” Current estimates tend to run between 19,000 and 22,000, but recent criticisms “underscore just how difficult it is to identify new genes, or even to define what a gene is” (Willyard 2018).
6. Lavelle 2009. Nucleoproteins are proteins bound up with DNA or RNA. A nucleoprotein complex would be a complex of DNA or RNA plus protein.
7. To get more specific about it, think of it this way. If, taking a double-stranded rope in hand, you insert a pencil between the strands and force it in one direction along the rope, you will find the strands winding ever more tightly ahead of the pencil’s motion and unwinding behind. An RNA polymerase, which must separate the two strands of DNA as it transcribes a gene, can in the right circumstances have an effect rather like the pencil: it will cause negative supercoiling (loosening of the double helix spiral) behind itself, and positive supercoiling ahead. And if, say, negative supercoiling has already occurred in the region being transcribed, the polymerase will find it much easier to separate the two strands and do its work. So in this way the variations in coiling along the length of a chromosome either encourage or discourage the transcription of particular genes.
8. To get a rough sense merely for the number of significant variations in DNA double helix conformation and the kind of effect they can have, here is a statement enumerating such variations and their bearing on a single regulatory feature, namely, the position of certain nucleosomes (referred to as “variant –1 nucleosomes,” which themselves play a key role in regulation of gene expression). There is no need to understand the different technical terms in order to get a feel for the complexity of the sculptural details of any particular stretch of DNA, and the kind of role these details can play in relation to gene expression.
Variant –1 nucleosomes [that is, nucleosomes at the places on DNA where gene transcription starts] exhibited a preference for sequences with altered features such as propeller twist, opening, electrostatic potential, minor groove width, rise, stagger, helix twist, and shear and roll. Variant –1 nucleosomes that shifted downstream in KDM5B-depleted ES [embryonic stem] cells preferred sequences with increased propeller twist, opening, electrostatic potential, stagger, minor groove width, rise, and buckle, while –1 variant nucleosomes that shifted upstream preferred sequences with decreased propeller twist, opening, electrostatic potential, stagger, minor groove width, rise, and buckle … Combined, these findings suggest that DNA shape predicts sequence preferences of canonical nucleosomes and variant nucleosomes. These results also suggest that histone DNA binding patterns such as bending or electrostatic interactions may be influenced by posttranslational modifications such as H3K4 methylation (Kurup, Campeanu and Kidder 2019).
9. Figure 3.3 credit: top image: LadyofHats (https://commons.wikimedia.org/wiki/File:Mitotic_Metaphase.svg), Public Domain via Wikimedia Commons. Bottom image: Afunguy (https://commons.wikimedia.org/wiki/File:Kinetochore.jpg), Public Domain via Wikimedia Commons.
10. The notion of “immaterial” causation is, of course, scarcely allowable in today’s science. Or so it is commonly thought. But this seems far from true. Is there not a sense in which every scientist implicitly agrees that ideas possess causal power? What about the idea of gravity — the ideal, form-giving aspect of it that we routinely formulate in mathematical thought? Isn’t this immaterial idea, or thought, definitively present in all analyses of our movements on earth?
The idea of gravity is, of course, a long way from the formative ideas we see at work in organisms. But no one has shown us inherent limits upon the kinds of ideas that might be embodied in the phenomena of the material world. In any case, just as we indisputably “see” the mathematics of gravity in earthly motions, we also and with equal persuasiveness “see,” for example, the striving for life evident in all organisms. Where physicists prefer to concern themselves with universal laws that apply to objects solely with regard to abstracted quantities such as mass, biologists deal with the behavior arising from within the qualitatively differentiated, more or less individuated “objects” (beings) of their science.
11. We are, of course, told that “desirable” and “undersirable” really refer to whether a trait is or is not conducive to an organism’s survival and therefore favored by natural selection. But ask yourself: How does this line of thought make more explicable what we have just heard about the activity of topoisomerases in the cell? To believe that every feature an organism actually possesses must be consistent with natural selection is no ground for saying that the materialistically conceived processes of natural selection can positively account for processes inexplicable in strictly physical terms. See the discussion of natural selection under “The shortest path to confusion is circular” in Chapter 18, (“Teleology and Evolution”).
Sources
Ball, Philip (2003). Portrait of a Molecule, Nature vol. 421 (January 23), pp. 421-2. doi:10.1038/nature01404
Dekker, Job and Tom Misteli (2015). Long-Range Chromatin Interactions, Cold Spring Harbor Perspectives in Biology 2015;7:a019356. doi:10.1101/cshperspect.a019356
Kouzine, Fedor, Suzanne Sanford, Zichrini Elisha-Feil, et al. (2008). The Functional Response of Upstream DNA to Dynamic Supercoiling in Vivo, Nature Structural and Molecular Biology vol. 15, no. 2 (February), pp. 146-54. doi:10.1038/nsmb.1372
Kurup, Jiji T., Ion J. Campeanu and Benjamin L. Kidder (2019). Contribution of H3K4 Demethylase KDM5B to Nucleosome Organization in Embryonic Stem Cells Revealed by Micrococcal Nuclease Sequencing, Epigenetics & Chromatin vol. 12, no. 20. doi:10.1186/s13072-019-0266-9
Lavelle, Christophe (2009). Forces and Torques in the Nucleus: Chromatin under Mechanical Constraints, Biochemistry and Cell Biology vol. 87, pp. 307-22. doi:10.1139/O08-123
Lewontin, R. C. (1992). The Dream of the Human Genome, New York Review (May 28), pp. 31-40. https://www.nybooks.com/articles/1992/05/28/the-dream-of-the-human-genome/
Lieberman-Aiden, Erez, Nynke L. van Berkum, Louise Williams et al. (2009). Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome, Science vol. 326 (October 9), pp. 289-93. doi:10.1126/science.1181369
Moss, Lenny (2003). What Genes Can’t Do. Cambridge MA: MIT Press.
Moss, Lenny (2012). Is the Philosophy of Mechanism Philosophy Enough? Studies in History and Philosophy of Science Part C: Studies in History and Philosophy of Biological and Biomedical Sciences vol. 43, no. 1 (March), pp. 164-72. doi:10.1016/j.shpsc.2011.05.015
Physorg (2014). Scientists Map the Human Loop-ome, Revealing a New Form of Genetic Regulation (December 11). https://phys.org/news/2014-12-scientists-human-loop-ome-revealing-genetic.html
Rao, Suhas S. P., Miriam H. Huntley, Neva C. Durand et al. (2014). A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping, Cell vol. 159, pp. 1665-80 (December 18). doi:10.1016/j.cell.2014.11.021
Talbott, Stephen L. (2010). The Unbearable Wholeness of Beings, The New Atlantis no. 29 (fall), pp. 27-51. Available at https://thenewatlantis.com/publications/the-unbearable-wholeness-of-beings Original version published in NetFuture no. 29 (fall), pp. 27-51. Also available at https://bwo.life/mqual/genome_5.htm
Talbott, Stephen L. (2024). How Do Biomolecules “Know” What To Do?, In Context #51 (spring). Latest version: https://bwo.life/bk/ps/biomolecules.htm
Wang, James C. (2009). Untangling the Double Helix. Cold Spring Harbor NY: Cold Spring Harbor Laboratory Press.
Willyard, Cassandra (2018). New Human Gene Tally Reignites Debate, Nature vol. 558 (June 21), pp. 354-55. doi:10.1038/d41586-018-05462-w
Zimmer, Carl (2015). Is Most of Our DNA Garbage? New York Times (March 5). https://www.nytimes.com/2015/03/08/magazine/is-most-of-our-dna-garbage.html
This document: https://bwo.life/bk/genes.htm
Steve Talbott :: What Brings Our Genome Alive?
