How Our Genes Come to Expression
Tags: chromosome/chromatin; epigenetics; holism; plasticity/genome; transcription; transcription/transcription factors
If your understanding of genetics comes from your newspaper’s science section, or a popular science magazine, or any other source intended for the general public, then you probably will not have been given the remotest glimpse of what actually goes on with the genes in our bodies. In fact, geneticists themselves have been known to lament how limited their knowledge of gene-related activity is, simply because the demands of professional specialization scarcely allow a wide field of view.
But it turns out that a wide field of view is the one critical prerequisite for any adequate understanding of genes. Only a broad survey can illustrate how every gene, like a significant word in a text, receives its full meaning only through the interweaving and converging influences issuing from all the elements of its context.
My aim here is to offer such a wider, “epigenetic” view — and to do so in the briefest space possible. If I succeed, you will begin to sense a biological landscape that reconfigures many long-standing assumptions, not only about genetics itself, but also about the character of all living activity.
High expectations: the
promise of molecular biology
After the discovery of the structure of the DNA double helix in 1953 and the elaboration of the “genetic code” during the early 1960s, the expression of a gene was thought of as the production of a functional protein corresponding precisely to instructions in the gene — coded instructions that were spelled out in the gene’s sequence of DNA “letters,” or nucleotide bases. The protein’s production, based on this sequence, was routinely described as a cut-and-dried, fully determined, rather mechanistic affair. The larger picture was sometimes summed up in this formula:
DNA makes RNA, RNA makes protein, and protein makes us.
A few key terms may help to flesh out the formula as it was then understood. (All the special vocabulary is elaborated in an online glossary at https://bwo.life/mqual/glossary.htm.)
The first step in gene expression was thought to be the binding of a protein transcription factor (one of many such factors existing in the cell) to DNA at or near a target gene. This led to the adjacent binding of a complex protein called “RNA polymerase” (often described as a “molecular machine”), which then transcribed the DNA sequence of the gene into an RNA molecule closely mirroring the DNA sequence.
Finally, the RNA was exported from the cell nucleus into the cytoplasm, where it was translated into a specific protein. The translation was carried out by another complex “molecular machine,” known as a ribosome. The sequence of amino acids in the resultant protein was said to have been coded for by the sequence of nucleotide bases in the gene and the similarly coded sequence of the RNA. A parallel was sometimes drawn with Morse code, in which a sequence of dots and dashes codes for a sequence of alphabetic letters.
The discovery of the entire scheme, so neat and tidy, testified to the impressive technical sophistication of the researchers, and was universally acclaimed.
But there was already a curiosity. Consider the picture. The production of a protein from DNA was initiated by a protein transcription factor. The “molecular machines” doing the work of transcription and translation consisted, in whole or in part, of proteins. Moreover, it was recognized that proteins were decisive for the very existence of DNA, as well as its replication, maintenance, and repair. So not only were proteins required in order to explain their own synthesis, but they were also required in order to explain the existence of DNA.1 At the same time, DNA was clearly required for the existence of proteins.
You might think the chicken-and-egg problem here would have given the scientific community pause during its single-minded, twentieth-century rush toward a gene-centered view of life. Was it really genes that made the organism, including its proteins? Or was it proteins that made the organism, including its genes? Or were both points of view terribly flawed and unbiological, so that we were being asked to rise to a more living and integral level of understanding where it is impossible to say that one thing unambiguously “causes” another?
Complications
Fast forward to today, and consider just one of the terms mentioned above: “transcription factor.” A riddle posed by many such protein factors involves their “promiscuous binding.” Transcription factors, of which there are over a thousand in the human genome, are not targeted to specific DNA sequences by some iron necessity. Most of them are quite capable of binding at thousands of locations throughout the genome — that is, at far more loci than they are actually found at in typical assays of living cells. In other words, we have to look for much more than a definitive, sequence-based targeting logic if we want to understand how transcription factors activate (or inhibit) specific genes in this or that specific kind of cell and context.
So the question arises, How does a transcription factor “know” which gene or genes to interact with? If its specificity — its ability to bind where it is needed — is not dictated by a simple and determinative match between its own binding domain and the DNA sequence it binds to, then how do we make sense of its well-directed activity? Is this activity merely expressing something like the logic at work in a humanly devised mechanism? Or is it more like a living language, where words can have diverse expressive potentials that are in part lent to them by their context?
The answer — or, rather, the many answers — are still unfolding today. The one indisputable truth is that it takes a molecular “village” — a vigorous and entire cellular context — to establish the correct and ever-changing relations between a transcription factor and the genes it helps bring to expression. The old idea that the relations among transcription factors, genes, and gene products are unambiguous — are governed by a fixed, necessary, and easily comprehended logic — is no longer tenable.2
Transcription factors and DNA engage in a complex play of form
To begin with, not just the DNA sequence, but also the moment-by-moment sculptural form, or conformation, of a DNA locus affects the binding potential of a transcription factor. This dynamically imposed form reflects the cellular environment. Also decisive are the plastic conformational potentials of the transcription factor itself. And then there are the many other essential molecules (”co-factors”) that may not even have the ability to bind to DNA, but which are nevertheless essential co-participants, along with transcription factors, in an interactive community through which a gene, or set of genes, is made ready for transcription.
For example, one way a transcription factor can contribute to the expression of a gene is by bending a short stretch of DNA into a shape conducing to further interaction. (For a striking, if highly schematic, illustration of this, see Figure 14.5 below.) By this means the initial presence of a transcription factor can make it easier than it would otherwise be for a second protein to bind nearby. In the case of one gene relating to the production of interferon (an important constituent of the immune system), “eight proteins modulate [DNA] binding site conformation and thereby stabilize cooperative assembly [of gene-regulating proteins]” (Moretti et al. 2008).
And so, despite the fact that “DNA is often mistakenly viewed as an inert lattice” onto which proteins bind in a sequence-specific way (Chaires 2008), the fact of the matter is altogether different. Proteins and DNA are caught up in a continual conversation of mutual influence and shifting form. It becomes obvious, then, that “No simple code combines all the various determinants of transcription factor binding specificity” (Slattery et al. 2014).
In other words, a transcription factor’s “recognition” of a DNA binding site is not a digital, yes-or-no matter, but a community judgment. And how could it be otherwise, given that no cell in our bodies (and no collection of molecules) lives merely for itself? Our activities always involve vast, cooperating communities of various sorts. Every cell and cellular organelle is caught up in a larger context of meaning and must be capable of adapting itself to, and supporting, virtually any of the infinitely varying activities we find ourselves engaging in.
A living flexibility is therefore crucial. So it is no surprise when one pair of researchers, studying a group of transcription factors in the genomes of animals, report “a dazzling array of strategies employed by [these] transcription factors to control gene expression.” The “emerging, unifying theme,” they say, is the ability of these transcription factors “to interact with many diverse partners. This high connectivity is probably crucial to assemble highly context-specific, transcriptionally active complexes at selected sites in the genome” (Bobola and Merabet 2017).
Genes and proteins interact in tangled causal webs
It is hard to take in the full significance of this “high connectivity,” which is typical of so many biological processes. One way to visualize the complications is to consider the fact that some transcription factors can target genes for other transcription factors. And, of course, this second group of transcription factors might target the genes for still other transcription factors as well as the genes or regulatory sequences associated with the first group. We can easily imagine the tangled causal webs resulting from this kind of inter-connectivity, where causal “arrows” can eventually circle back to their starting point. Unsurprisingly, there are entire fields of research today given over to complex gene and regulatory networks such as shown in Figure 14.1.
Figure 14.1. Transcription regulation network of Parkinson’s disease, showing differentially expressed genes (pink) together with some of the transcription factors (blue) playing a role in regulating those genes. The figure is too small to read — purposely. Researchers sometimes lightheartedly refer to such diagrams as “hairballs,” which is about all you need to know.3
Returning to the puzzle of transcription factor “promiscuity”: this word reflects neither undisciplined profligacy nor uncertainty of function. Rather, it points to the unbounded, context-specific potentials of transcription factors. Their contribution to essential cellular processes, after all, is properly focused and far from promiscuous. They are caught up within a wisdom that seems to “know” exactly what it is doing. It’s just that this doing is complex and living — flexible and adaptive — far beyond what a simple, definitive, one-dimensional mapping between DNA sequence and a rigidly complementary protein shape would allow. This flexibility is what allows community-tuned activity in the larger surround to influence local goings-on in endlessly nuanced ways — all so as to satisfy the needs of the current context.
It is important to underscore here a fact we have found ourselves coming up against throughout this book: the tangled causal web we discover in organisms is not merely a matter of complexity. There are many nonliving physical contexts so complex that, as a practical matter, we cannot easily trace precise lines of cause and effect. This is true of eddies in a great river or in the atmosphere, and it is even true of some kinds of computer program. And yet no one would doubt in these cases that the relevant causes could be traced, at least in principle, or that the tracing would give us what is usually (if erroneously) considered to be a full accounting of what we are looking at.
But, as I began explaining in Chapter 2, the purposive behaviors of organisms exhibit a kind of coherence and meaning that is not satisfactorily explained when we look only at principles of physical causation. The “causal confusion” in the organism’s case is not due merely to the complexity of the always lawful and harmonious physical relations, but rather to the fact that purposive and narrative explanation must be found at a “higher” level of meaning than physical lawfulness. The significance of what is going on is recognized only when we consider the insistent coordinating principles through which physical events are caught up in serving the needs and interests of organisms. Because concepts such as “need” and “interest” are incommensurable with the accepted principles of physical explanation, they demand recognition as explanatory principles in their own right.
The cell holds DNA in an intimate and instructive embrace
Our brief discussion of genes and transcription factors has, so far, been hopelessly simplistic. The chromosomes in our cells do not consist of a naked DNA double helix sporadically bound at particular sequences by this or that transcription factor. The picture is wholly different. Our DNA is intimately bound up with a massive, intricate, and dynamic protein-RNA-small molecule complex that, together with the DNA, is called chromatin. “Chromatin,” in other words, can pass as simply a name for the full substance of chromosomes. The proteins in this complex are as weighty as the DNA itself — and much more active and directive when it comes to gene expression.
Some of the protein constituents of this chromosomal substance — both the longer-term and the many transient constituents — can bind directly to DNA, thereby facilitating, blocking, or modifying the transcription of this or that gene. But other elements of chromatin, while not directly bound to DNA, nevertheless contribute crucially to the regulation of gene expression. Overall, the molecular factors associated with chromatin play roles such as the following:
- they help to condense or decondense the packing of the DNA (more tightly condensed DNA tends to be less accessible to activating factors);
- they move chromosomes or parts of chromosomes to different regions of the cell nucleus (the interior of the nucleus tends to be more transcriptionally active than the periphery);
- they attach parts of chromosomes to the nuclear envelope (many factors at or near the envelope bear on gene expression);
- they interweave and (almost miraculously, it might seem) disentangle chromosomes, while also forming decisively important chromosome loops (such as those we heard about in Chapter 3, “What Brings Our Genome Alive?”) — all so as to form various-sized “communities” of functionally related chromosomal loci;
- they untwist (loosen) the two strands of the double helix in some places and twist them more tightly in others, which can make the difference between a gene’s accessibility or inaccessibility to transcription factors;
- and they alter the electrical characteristics of particular loci (yet another feature bearing on the expression of affected genes).
As you may surmise, then, it’s not as if the power to determine gene expression outcomes is one-sidedly delegated to any genetic sequences, any transcription factors, or any other entities. It is rather as if the result arises in the way a musical performance is evoked from a jazz ensemble. A distinct locus of DNA certainly offers its own expressive potentials, but there is no telling — no predicting solely from an analysis of the sketchy DNA “musical score” — how the locus may be employed within the improvised, multi-cellular performance leading from a single fertilized egg cell to the mature human being.
But perhaps we would do better to imagine an exquisitely detailed, never-ending, self-assured, yet highly improvisational dance involving billions of molecular dancers within a cell — all coordinated with the choreography in neighboring cells and with the ongoing story of the organism as a whole. The performance, involving the fluid identity of countless players, is a long way from that of calculating or information-processing hardware and software.
In any case, the present point is that our DNA is thoroughly “wedded” to — bound together with — an almost unfathomably intricate arrangement of protein, RNA,4 and small molecules. The protein and RNA constituents of this chromatin complex are fully as “information-rich” as the DNA. Genes, as such, cannot do anything, and certainly cannot transcribe themselves. The information-rich, if unquantiable, doing is in large part a function of the associated proteins, which, among other things, thereby participate in their own genesis. Alongside them are many other molecules, including water molecules (Chapter 5, “Our Bodies Are Formed Streams”), all of whom give collective expression to the purposive coherence of the cell as a whole.
I have so far offered only a rather vague and general description of the highly effective embrace in which DNA is held. In later sections we will look further at some of its key features.
Getting started
is hard to do
Meanwhile, leaping tall edifices of thought in a single bound, we will pass over the question how cells “know” which genes need to be expressed within the current context of a person’s activity and within the trillions of cells constituting our bodies. We will also avoid asking how any single cell — which can play only a spatially minute part within an organ such as the liver or within a process such as wound healing — finds its own proper role in whatever the current larger performance happens to be. And so, assuming all the necessary contextualization and direction to be somehow wisely taken care of,5 we will imagine just one cell embarking on a single task: to give expression to one among its 20,000 or so genes. How might this cell proceed?
Our imaginative exercise will necessarily be more than a little artificial. That’s because we need to think one thing at a time, whereas in the cell countless mutually entangled things are all happening at once. But we will try to make the best of it.
You may recall from Chapter 3 (“What Brings Our Genome Alive?”) that packing DNA into a typical human cell nucleus is like packing about 24 miles of very thin, double-stranded string into a tennis ball, with the string divided into 46 separate pieces, corresponding to our 46 chromosomes.
To locate a modest-sized protein-coding gene within all that DNA is like homing in on a half-inch stretch within those 24 miles.6 Or, rather, two relevant half-inch stretches located on different pieces of string, since most of our cells have two copies of any given gene, residing on different chromosomes. Except that sometimes one copy differs from the other and one version is not supposed to be expressed, or one version needs to be expressed more than the other, or the product of one needs to be modified relative to the other. So part of the job may be to distinguish one of those half-inch stretches from the other, and to act differently in the two cases. “Decisions” everywhere, it seems.
As a functional unit, a gene must participate in a performance appropriate to the current cellular and extra-cellular context, and the highly distributed activity responsible for its function must be cobbled together by the cell according to the needs of the moment. There is no predefined path to follow once the cell has located the “right” half inch or so of “string,” or once it has done whatever is necessary to bring that locus into proper relation with other chromosomal loci participating in, and essential to, a joint performance.
One issue has to do with the fact that there are two strands of the double helix, and (in a chemical sense) these complementary strands “point” in opposite directions. In humans, protein-coding sequences can occur on both strands. Likewise, transcription (of both protein-coding and regulatory sequences) occurs on both strands, which is to say that the transcribing enzyme (RNA polymerase) can move in either direction along the double helix. The direction chosen — that is, the strand along which the RNA polymerase will move — depends on the meaning within the current context of the sequences that exist at the current locus. Somehow, acting within and guided by its present context, RNA polymerase must have the “good sense” to choose the appropriate activity from among the various possibilities.
And even when the cell “knows” to initiate transcription in one particular direction, it must “choose” the exact point in the genetic sequence at which to begin. Different starting points can yield functionally distinct results. “Many studies focusing on single genes have shown that the choice of a specific transcription start site has critical roles during development and cell differentiation, and aberrations in … transcription start site use lead to various diseases including cancer, neuropsychiatric disorders, and developmental disorders” (Klerk and ’t Hoen 2015).
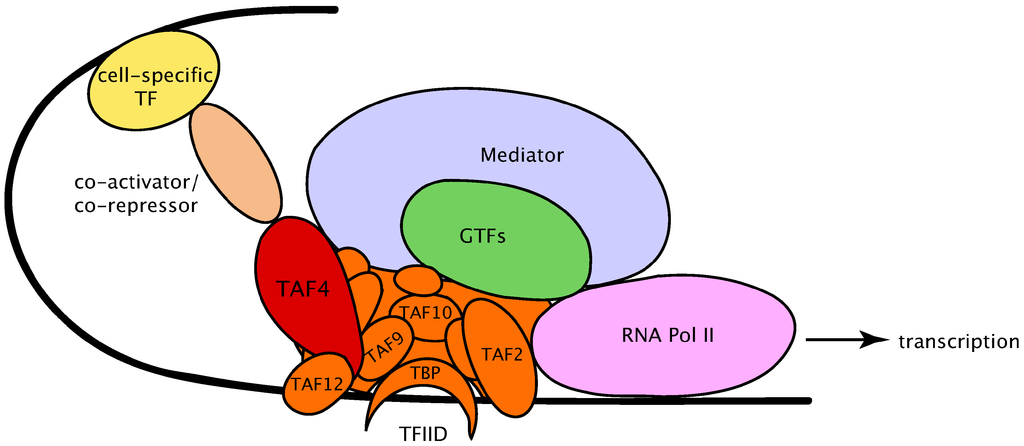
Figure 14.2. The pre-initiation complex (cartoon representation).7
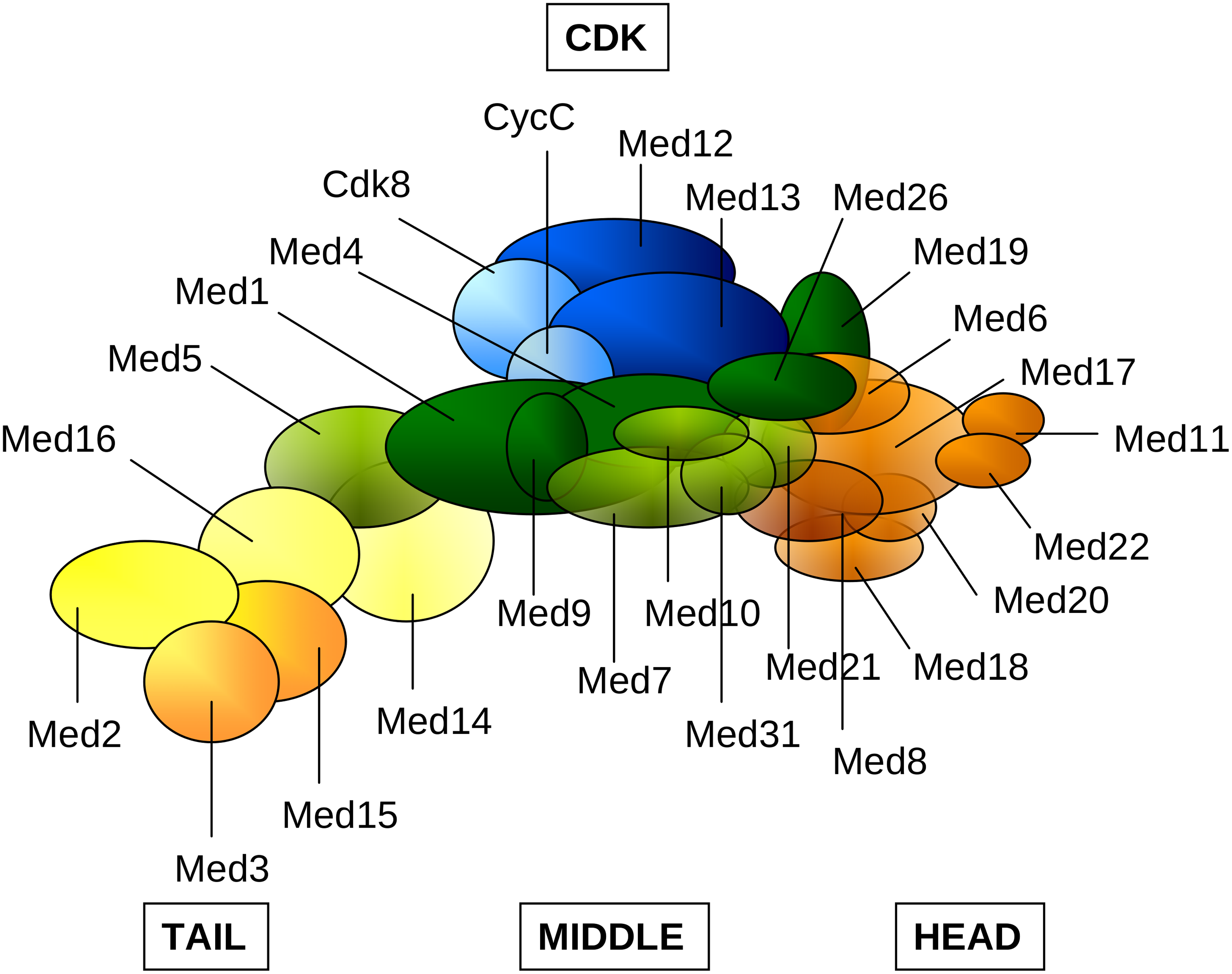
Figure 14.3. Some subunits of the Mediator complex (cartoon representation) captured at the CDK gene locus.8
Intertwined with all the preceding issues is the cell’s task of assembling a pre-initiation complex (PIC). This variable arrangement of regulatory elements typically sets the stage for the transcriptional activity to follow. Figure 14.2 is a cartoon figure that merely names some of the protein PIC constituents that arrange themselves on DNA (shown as a black line) near locations where gene transcription is to begin. You needn’t concern yourself with names and meanings, beyond the general description I am offering now.
The cell’s narrative at this point could hardly be more dramatic — or more subtle. The largest oval in Figure 14.2, named “Mediator,” is a massive molecule consisting (in humans) of 26 protein subunits (Figure 14.3) arranged in modules and interacting in numerous ways among themselves, as well as with other PIC constituents and “visiting” molecules. Depending on context, Mediator can vary endlessly in both subunit composition and function. Its effects upon gene expression are many, and still only fragmentarily grasped.9
Figure 14.4 shows the known interaction partners for the Mediator subunits in just one cell type — mouse neural stem cells. The figure omits the numerous interactions among the Mediator subunits themselves. It also omits the interactions among the molecules shown in the surrounding circle. And, perhaps most importantly, it omits the interactions those molecules have with still others not shown in the diagram. For it is just a fact that each of these molecules shown in the outer circle could be made the center of its own diagram. Reflecting on this can usefully remind us of what it means to say that all biological activity in a cell, no matter how micro-focused our vision, turns out upon broader inspection to be an almost impossibly intricate and coordinated activity of the whole.
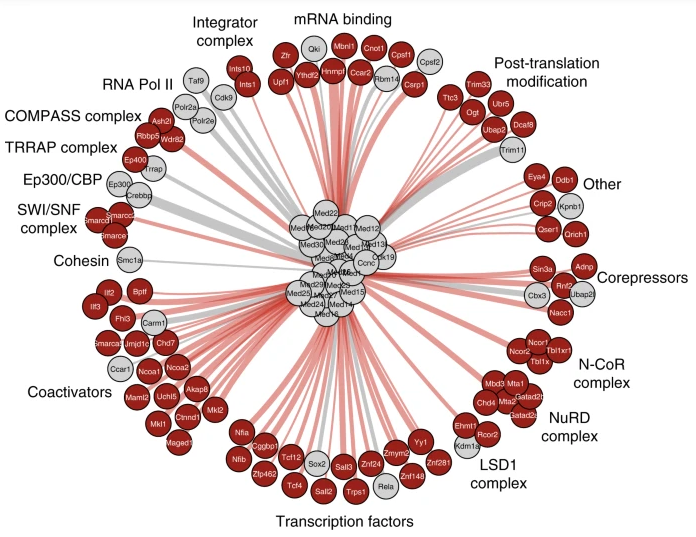
Figure 14.4. Interactions of the Mediator complex in mouse neural stem cells. Mediator subunits are shown in the middle. Gray circles and lines represent interaction partners that were already known at the time (2019) when the research was carried out. Red circles represent interactions newly discovered by the authors of the paper from which this figure was taken.10
And, of course, Mediator is just one element of the PIC. Each of the other elements has its own story to tell. The entire PIC was once regarded as a rather mechanical, routine, and mostly unvarying assembly of “parts” whose unproblematic duty was to initiate gene transcription in a standard way. But, of course, that was to overlook how thoroughly every aspect of gene expression must vary if it is to serve the needs of a living being. The PIC is now seen to be an infinitely modifiable, highly dynamic complex, responding both to the immediate DNA context and to influences arriving from distant reaches of the cell. Its overall “decision-making” role, which can differ from one gene to the next, is hardly the functioning of a routinely analyzable mechanism.
It doesn’t require of the reader a technical penetration of these figures to get a sense for the kind of thing that is going on — especially if one keeps in mind that we are talking, not about rigid machinery of the sort we are familiar with in our daily lives, but rather about molecular interactions within a highly fluid context where machine-like constraints to forcibly channel the interactions are altogether absent.
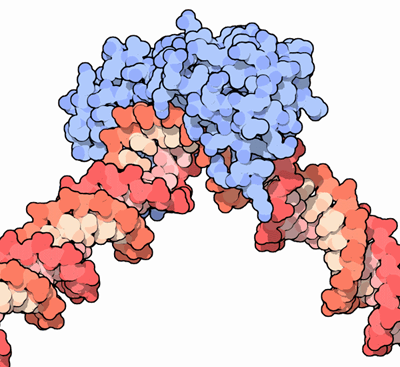
Figure 14.5. DNA (red) in the grip of the tata-binding protein (blue).11
I will mention here just one other element of the pre-initiation complex. Figure 14.5 shows DNA (in a wholly artificial, simplistic, and impossibly rigid, concrete representation) being “gripped” by the tata-binding protein (TBP), shown in blue. (TBP is also seen as the crescent-moon shape at the bottom of Figure 14.2.) The protein “clasps” the DNA in an intimate and rather tortuous manner — a clasp that might remind one of the forcible interaction between two human wrestlers.12 A severe bend of about eighty degrees is thereby applied to the double helix. This bend, which also tends to pull the two strands of the helix apart, is a general prerequisite for the assembly and activity of the rest of the PIC. As always, the cell is doing something sculptural, not narrowly informational in the usual sense.
Carrying on
As we heard at the outset, the (protein) enzyme that transcribes DNA into RNA is RNA polymerase.13 The enzyme certainly does not work alone, however, and its task is by no means automatic. To begin with, its critical interactions with various elements of the pre-initiation complex help determine whether and exactly where transcription will begin. Then, after those “decisions” have been made, RNA polymerase moves along the double helix transcribing the sequence of genetic “letters” into the complementary sequence of an RNA.
Throughout this productive journey, which is called elongation, the RNA polymerase still keeps good and necessary company. Certain molecular co-activators modify it during its transit of a gene’s sequence, and these modifications not only enable transcription elongation to begin, but also provide binding sites for yet other proteins that will cooperate throughout the transcription journey. The collective interaction here, as in the activities discussed above, can vary in many details from one context to another — all in order to contribute to a meaningful narrative that could hardly repeat itself in exactly the same way.
The table below offers some perspective on the number and variety of protein factors influencing elongation. You need not puzzle over the details. A quick browse of this incomplete listing (as of 2013) will give you at least an inkling of the kind of intricate complexity the cell must organize in order to carry out transcriptional elongation. As always, it is important to realize that each of the factors listed here enters the picture out of its own world of regulation. At the molecular level of the organism we are always looking at ever-widening circles of interaction, without limit. It’s just a question of how narrowly we choose to focus our attention — and how much of the context we consequently block from view.
Table 14.1. Don’t Read This Table! (Just feel it.) Some factors regulating RNA polymerase elongation (copied from Kwak and Lis 2013).
| Class | Factor name | Function | Related factors and notes |
|---|---|---|---|
| GAGA factor | GAF | Generates nucleosome-free region and promoter structure for pausing | NURF |
| General Transcription Factors | TFIID | Generates promoter structure for pausing | |
| TFIIF | Increases elongation rate | Near promoters | |
| TFIIS | Rescues backtracked RNA polymerase II | RNA polymerase III | |
| Pausing factors | NELF | Stabilizes RNA polymerase II pausing | |
| DSIF | Stabilizes RNA polymerase II pausing and facilitates elongation | ||
| Positive elongation factor | P-TEFb | Phosphorylates NELF, DSIF, and RNA polymerase II CTD for pause release | |
| Processivity factors | Elongin | Increases elongation rate | |
| ELL | Increases elongation rate | AFF4 | |
| SEC | Contains P-TEFb and ELL | Mediator, PAF | |
| Activator | c-Myc | Directly recruits P-TEFb | |
| NF-κB | Directly recruits P-TEFb | ||
| Coactivator | BRD4 | Recruits P-TEFb | |
| Mediator | Recruits P-TEFb via SEC | ||
| Capping machinery | CE | Facilitates P-TEFb recruitment, counters NELF/DSIF | |
| RNMT | Methylates RNA 5’ end to complete capping | Myc | |
| Premature termination factors | DCP2 | Decaps nascent RNA for XRN2 digestion | Dcp1a/Edc3 |
| Microprocessor | Cleaves hairpin structure for XRN2 digestion | Tat, Senx | |
| XRN2 | Torpedoes RNA polymerase II with RNA 5’-3’ exonucleation | ||
| TTF2 | Releases RNA polymerase II from DNA | ||
| Gdown1 | GDOWN1 | Antitermination and stabilizes paused RNA polymerase II | TFIIF, Mediator |
| Histone chaperone | FACT | H2A-H2B eviction and chaperone | Tracks with RNA polymerase II |
| NAP1 | H2A-H2B chaperone | RSC, CHD | |
| SPT6 | H3-H4 chaperone | Tracks with RNA polymerase II | |
| ASF1 | H3-H4 chaperone | H3K56ac | |
| Chromatin remodeler | RSC | SWI/SNF remodeling in gene body | H3K14ac |
| CHD1 | Maintains gene body nucleosome organization | FACT, DSIF | |
| NURF | ISWI remodeling at promoter | GAGA factor | |
| Poly(ADP-ribose) polymerase | PARP | Transcription independent nucleosome loss | Tip60 |
| Polymerase-associated factor complex | PAF | Loading dock for elongation factors | SEC, FACT |
| Histone tail modifiers | MOF | Acetylates H4K16 and recruits Brd4 | H3S10ph, 14-3-3 |
| TIP60 | Acetylates H2AK5 and activates PARP | ||
| Elongator | Acetylates H3 and facilitates nucleosomal elongation | Also in cytoplasm | |
| Rpd3C (Eaf3) | Deacetylates and inhibits spurious initiation in gene body | H3K36me3 | |
| SET1 | Methylates H3K4 | MLL/COMPASS | |
| SET2 | Methylates H3K36 and regulates acetylation-deacetylation cycle | Rpd3C | |
| PIM1 | Phosphorylates H3S10 and recruits 14-3-3 and MOF | ||
| RNF20/40 | Monoubiquitinates H2BK123 and facilitates nucleosomal DNA unwrapping | UbcH6, PAF |
I will mention here only one aspect of this cooperation of multiple factors. Transcription is an essentially rhythmical performance, with various sorts of pauses along the way. (Again, dynamic sculpture, or dance!) One pause of great significance occurs after RNA polymerase has just begun transcribing DNA but before it has fully separated from the pre-initiation complex. The factors that influence whether transcription will continue at this point — or remain paused for an extended period — play a large role in the regulation of gene expression.
But once that first pause is ended, the elongation journey often continues to be marked by a series of further, generally briefer pauses. These have to do, at least in part, with the need to disengage DNA from its intimate mutual embrace with certain constituents of chromatin (histone complexes, about which we will learn more below). The polymerase has various assistants to aid in this disengagement, which may involve disassembly of the protein complexes. Typical of chromatin in general, these histone complexes are rich repositories of regulatory information, so they will need to be reassembled behind the transcribing complex, and the remarkably nuanced meanings embodied in their composition and structure will somehow have to be preserved, reestablished, or modified.
So the rhythm of pauses depends, at least in part, on the polymerase’s helper molecules and on the positioning of certain protein complexes along the double helix, both of which will vary from one gene to another and even from one time to another. All this, and not just the so-called genetic code as such, shapes the functional significance of the DNA sequence within its chromosomal context. As we will see shortly, different versions of a protein may be produced, depending on the timing of the pauses.
Shaping a
significant end
Finally — and mirroring all the possibilities surrounding initiation of gene transcription — there are the issues relating to its termination. Again, they are far too many to mention here. Transcription may conclude at a more or less canonical terminus, or at an alternative terminus, or it may proceed altogether past the gene locus, even to the point of overlapping what, by usual definitions, would be regarded as a separate gene farther “downstream.” The cell has great flexibility in determining what, on any given occasion, counts as a gene, or transcriptional unit.
The very last part of the transcribed gene is generally non-protein-coding, but nevertheless contains great significance. Examining this region in a single gene, one research team identified “at least 35 discrete regulatory elements” to which other molecules can bind (Kristjánsdóttir, Fogarty and Grimson 2015). Importantly: additional dramatic and diverse regulatory potentials arise from the customized “tail” that the cell commonly adds to the end of an mRNA after its transcription from DNA. The regulatory processes called into play by this tail can affect everything from the stability of the mRNA to its cellular localization and the efficiency of its translation into protein. It can even play a role in determining exactly what protein will ultimately be produced. And the patterns of these added tails tend strongly to differ from one tissue type to another. “Decisions” yet again.
Much of this post-transcriptional regulation is accomplished by proteins and other molecules that bind, not only to the end, but also to the various regulatory sequences at the head of the RNA transcript. It all occurs in a context-sensitive manner, where cell and tissue type, phase of the cell cycle, developmental stage, location of the transcript within the cell, and converging environmental factors, both intra- and extra-cellular, may all play a role.
But it’s not only the RNA sequence that provides opportunities for management by the cell. The three-dimensional, folded structure of the RNA molecule offers boundless occasion for further regulation. So here, as with DNA, we find gene expression to be in part a matter of sculptural performance. And, again, it is not just a matter of static form, but of movement. According to molecular biologists at the University of Michigan and Duke University, “RNA dynamics play a fundamental role in many cellular functions”:
[There are] many structural maneuvers that occur over timescales ranging from picoseconds to seconds … These transitions include large-scale secondary-structural transitions at [greater than tenth-of-a-second] timescales, base pair/tertiary dynamics at microsecond-to-millisecond timescales, stacking dynamics at timescales ranging from nanoseconds to microseconds, and other “jittering” motions at timescales ranging from picoseconds to nanoseconds. RNAs often harness multiple modes to achieve complex "functionality" (Mustoe et al. 2014).
From genetics
to epigenetics
“Epigenetics” refers to that which is not genetics as such, but rather is “added to,” or “on top of” genetics. You might therefore think that the transcription factors, RNA polymerases, and other proteins mentioned above, which are not themselves genetic elements, would be treated under the heading of epigenetics. Oddly, however, this has not been the case. Presumably, the reason is that these factors have for so long been taken for granted as if they were mere adjuncts to the “controlling logic” of DNA sequences.
But this never made much sense. What I have tried to suggest in my descriptions above is that these “mere tools” are more and more being recognized as participants in a dynamic communal context out of which alone our genes come to disciplined expression according to the needs of each cell.
Now, however, it is time to approach — albeit with painful brevity — what is generally considered the epigenetic mainstream. After all, we now know that gene transcription is merely a small part of all the activity shaping gene expression. The many processes “on top of” transcription are fully as rich and multifaceted as the various features of transcription itself.
We have already heard about RNA splicing, which we looked at in Chapter 8, “The Mystery of an Unexpected Coherence.” As we learned in that chapter, cells don’t just passively accept the RNAs that emerge from the transcription process, but rather “snip” them into pieces and “stitch” (splice) some of the pieces back together, while leaving others aside for purposes both known and unknown. It happens that these operations typically begin before the RNA is fully transcribed, and the rhythm of pauses by RNA polymerase during elongation influences which pieces are chosen for the mature transcript.
For the vast majority of human genes the splicing operation can be performed in different ways, yielding distinct protein variants (often called isoforms) from a single RNA. It would be hard to find any major aspect of human development, disease etiology, or normal functioning that is not dependent in one way or another on the effectiveness of this liberty the cell takes with the products of its gene sequences.
But RNA splicing is hardly the end of it. Through RNA editing the cell can add, delete, or substitute individual “letters” of the RNA sequence.14 Or, leaving the letters in place, the cell can apply over 170 distinct chemical modifications to them.15 Both the editing and the modifying are major topics in themselves, but not ones we can linger on here.
MicroRNAs: a large world of tiny regulatory factors
An entire, diversified area of research involves small, non-protein-coding RNAs. There are many different kinds of noncoding RNAs, but the only ones we will discuss here are known as microRNAs (miRNAs), which are generally derived through the cleaving and processing of longer RNAs. A microRNA commonly joins forces with a large protein complex, called the RNA-induced silencing complex (RISC). The microRNA guides the RISC to specific mRNAs by means of (sometimes only rough) base pair complementation. (See “base pair complementarity” in the online glossary at https://bwo.life/mqual/glossary.htm#base_pair.) Once a target mRNA is located, the RISC can cleave or otherwise degrade it, or else block its translation. In this way a typical microRNA can degrade or tune the amounts of a considerable number of different mRNAs.
Such degradation is an example of RNA decay in general, for which there are many different, interwoven pathways in cells. It is easy to overlook the fact that decay is fully as important — and fully as much in need of careful regulation — as the production of the RNA in the first place. During development, for example, cell differentiation would be impossible if the RNAs and proteins appropriate for an earlier form of a cell could not be recycled. In this way their constituent nucleotides or amino acids can support synthesis of new RNAs and proteins necessary for the cell’s forthcoming, more differentiated form. Such a refocusing of energies may be required by any changing conditions that require fresh responses from the cell.
MicroRNAs are key fine-tuners of the relative numbers of mRNAs in a cell under any given circumstances — and therefore also of the relative numbers of various proteins. We can only wonder how the microRNAs are “instructed” by the larger context so as to “know” what these relative numbers ought to be. But we do know some of the means employed.
One of the current stories about the role of microRNAs in regulating gene expression points to a complexity almost beyond all hope of detailed understanding. Evidence suggests that just about any RNA in the human body can help to regulate any number of other RNAs, just as it in turn is regulated by them. This intertwining of fates is due not only to the competition for resources (an extremely abundant RNA, by monopolizing the available amino acids in a cell, can make it more difficult for other RNAs to be translated into protein), but also to the impact of microRNAs. Here’s one way it works:
Many protein-coding RNAs are densely covered with binding sequences for microRNAs, so that a typical microRNA will find about 200 different RNA species it can target for decay or modification. This means that if a particular RNA is being highly expressed — and all the more if it is a “microRNA sponge” possessing multiple binding sites for a specific microRNA — it can have the effect of up-regulating other RNAs that are targets for the same microRNA. It “soaks up” most of the microRNAs that might otherwise degrade those targets. The RNAs that in this way regulate other RNAs by competing for shared microRNAs are known as “competing endogenous RNAs” (ceRNAs).
One research group (Tay, Rinn and Pandolfi 2014) traced the relations among a small network of twelve ceRNAs, which included the RNAs, PTEN (derived from the PTEN gene) and PTENP1 (derived from the PTENP1 gene). PTEN, when translated, yields a protein that is, among other things, a tumor suppressor. (It also appears to facilitate cell migration, and to play a part in the adhesion of cells to each other.) PTENP1, on the other hand, is an RNA derived from a so-called “pseudogene,” assumed to result evolutionarily from a mutational duplication of the PTEN gene, followed by further mutations compromising its protein-coding function. Pseudogenes are one more example of those many DNA elements, once written off as nonfunctional “junk,” which are now being “caught in the act” playing important roles.
In the present case, we know at least one role for PTENP1. Its RNA may be incapable of being translated into protein, but it nevertheless shares many microRNA binding sites with the PTEN RNA. By sequestering those microRNAs away from PTEN, PTENP1 allows the tumor-suppressor to be expressed at proper levels. If, on the other hand, the pseudogene becomes dysregulated for some reason so that PTENP1 is not produced, then microRNAs that would otherwise bind to PTENP1, end up instead binding to, and repressing, PTEN, which reduces its tumor-suppressing activity. It has in fact been shown that PTENP1 functioning is selectively lost in certain human cancers, consistent with its importance as a microRNA sponge.16
And yet, the situation is actually much “worse” than I have so far indicated. MicroRNAs can also regulate other microRNAs, whether by direct targeting or, indirectly, by targeting transcription factors or regulators of those other microRNAs. For example, one particular microRNA (known as miR-499) was shown not only to regulate target genes (via their mRNAs) in the usual way, but also altered the expression of 11 other miRNAs. These changes resulted in 969 down-regulated genes, only 7.8 percent of which were directly targeted by miR-499. In other words, “hundreds of genes may be altered in expression” via these indirect pathways radiating from a single microRNA (Hill and Tran 2021).
Here we see the same obstacle to any straightfoward causal understanding that we encountered above regarding transcription factors activating or repressing other transcription factors. Tracking the mutual, broad-scale, and often subtle interactions where “everything seems to be affecting everything else” will presumably challenge researchers for a very long while. It looks like a classic picture of the unanalyzable holism of all cellular processes. All the other interwoven aspects of gene regulation discussed in this chapter, when added together, only further complicate the problem of unanalyzability.17
DNA methylation
Some epigenetic processes profoundly implicated in gene expression transform the DNA sequence itself. That is, they modify the nucleotide bases (“letters”) of the so-called “genetic code.” One of these processes, known as DNA methylation, is extremely important for gene regulation.
DNA methylation is the addition of a methyl group (with chemical formula –CH3) to certain DNA bases. There are four different bases in DNA, and the one most commonly methylated is cytosine. In its methylated form, this has been referred to as the “fifth base of DNA.” Millions of bases throughout the genome are selectively and dynamically methylated in the cells of normal human tissues. The difference between a methylated and unmethylated base is hardly less significant, in its own way, than the difference between one base and another. But, unlike the general rule for the “raw” sequence of DNA bases, the methylation of those bases can be altered during development and in response to environmental influences. In this sense, much of our DNA inheritance is not at all the fixed-once-and-for-all destiny it is so often taken to be.
An “attached” methyl group is said to “tag” or "mark" the affected base. However, words such as “attach,” “tag,” and “mark” are grossly inadequate, suggesting little more than an annotation in the margin of a text, or a digital label on an otherwise unchanged entity. But in fact what DNA methylation gives us is chemical transformation — the metamorphosis of many millions of letters of the human genome under the influence of pervasive and incompletely understood cellular processes. And the altered balance of forces — the modulation of chemical, electrical, and sculptural qualities of chromosomes — resulting from all these chemically transformed bases, certainly plays with endless possible nuances into the expression of our genes.
We have been learning about the extreme consequences of these metamorphoses. In the first place, the transformations of structure brought about by methylation can render DNA locations no longer accessible to the protein transcription factors that might otherwise bind to them in order to activate nearby genes. On the other hand, by changing the local physical properties of the double helix, methylation “is observed to either inhibit or facilitate [DNA] strand separation, depending on methylation level and sequence context” (Severin et al. 2011). This has a direct effect on gene expression — for example, because strand separation is essential for the work of the polymerase that transcribes DNA.
Many proteins that recognize and bind specifically to methylated sites are then able to recruit other proteins that restructure and functionally alter the chromatin — for example, condensing it in a manner conducing to gene repression throughout an entire chromosomal region.
It would be difficult to overstate the pervasive role of this epigenetic factor in the organism. Stephen Baylin, a geneticist at Johns Hopkins School of Medicine, says that the silencing, via DNA methylation, of tumor suppressor genes is “probably playing a fundamental role in the onset and progression of cancer. Every cancer that’s been examined so far, that I’m aware of, has this [pattern of] methylation” (quoted in Brown 2008). In one study among various others — a study of colorectal cancer tissues — the researchers identified 1549 genomic regions with methylation patterns differing from the patterns in similar, non-cancerous tissues (Wei et al. 2016). There are often many more methylation anomalies in cancerous tissues than there are mutated genes.
In an altogether different vein, researchers have found that “DNA methylation is dynamically regulated in the adult human nervous system.” Distinctive patterns of DNA methylation are associated with Rett syndrome (a form of autism) and various kinds of mental retardation. Changing patterns of methylation also figure in aging, and constitute a “crucial step” in memory formation (Miller and Sweatt 2007).
Among many other things, DNA methylation appears to play a key role in tissue differentiation; in the activation (rather than only the repression) of gene transcription; and in the regulation of alternative RNA splicing. And, as by now we might expect, DNA methylation itself is regulated by processes converging from all corners of the cell and larger context.
The nucleosome: a
complex marriage
of DNA and protein
Nothing more vividly illustrates the cell’s dynamic and transformational “embrace” of its DNA than the thirty million or so nucleosomes that form the main bulk of human chromosomes. Each nucleosome consists of several histone proteins complexed together in a core particle, around which various other proteins help to bend and wrap the rather stiff DNA double helix. The DNA circles the core particle approximately twice and is (more or less) held in place there, largely by means of electrostatic forces and hydrogen bonding. It is time to focus on this remarkable protein-DNA complex — a complex that, for all its centrality, scarcely figures in the broader public understanding of genetics.
Figure 14.6 is an electron microscope-derived image obtained in the 1970s by the discoverers of the nucleosome, Ada and Donald Olin, who were then researchers at the University of Tennessee and Oak Ridge National Laboratory. You can see the nucleosomes as “beads” along the string-like DNA.
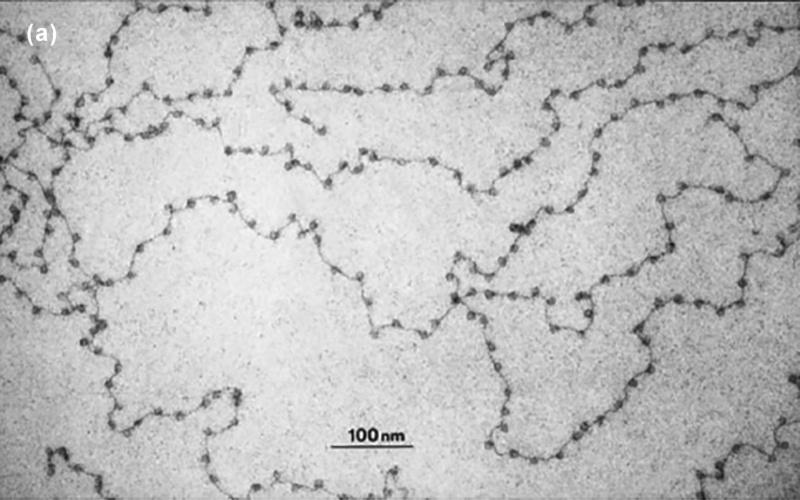
Figure 14.6. DNA (black “string”) and nucleosomes (“beads” on the string), as imaged by an electron microscope.18
A nucleosome most commonly consists of eight histone proteins (two copies of each of four histones, known as H2A, H2B, H3, and H4). The two stretches of linker DNA at the entry and exit points of the nucleosome are often held together by a linker histone (H1). The latter plays a role both in influencing how the DNA is bound to the core particle, and also in managing the packing together of neighboring nucleosomes.19 (See the cartoon representation in Figure 14.7.)
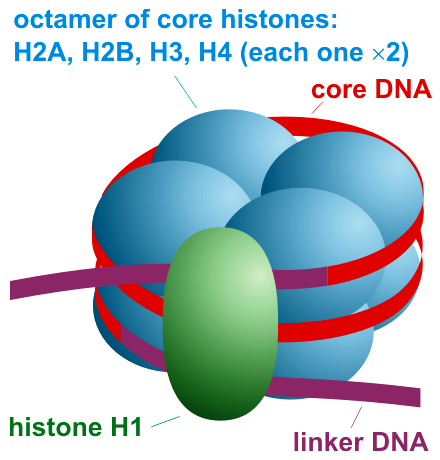
Figure 14.7. A schematic representation of a nucleosome, together with the linker histone (H1) and the encircling DNA.20
I referred earlier to the challenge of packing all the DNA of a cell into the space of the nucleus. As it happens, nucleosomes play a large role in this packing. Depending on their arrangement, which varies with the context, they help to organize the DNA molecule into a fiber that is said to be anywhere from (roughly) 1/5 to 1/50 of the uncondensed length. Something like 75 percent of our genome is wrapped up in nucleosomes, and a typical gene will have scores of nucleosomes within its body. This radically alters the popular image of a chromosome as a vast, uninterrupted length of the spiraling double helix.
Figure 14.8 shows (again in cartoon form) nucleosomes with and without linker histones, as well as the varying degrees of DNA compaction that can be achieved with the aid of nucleosomes.
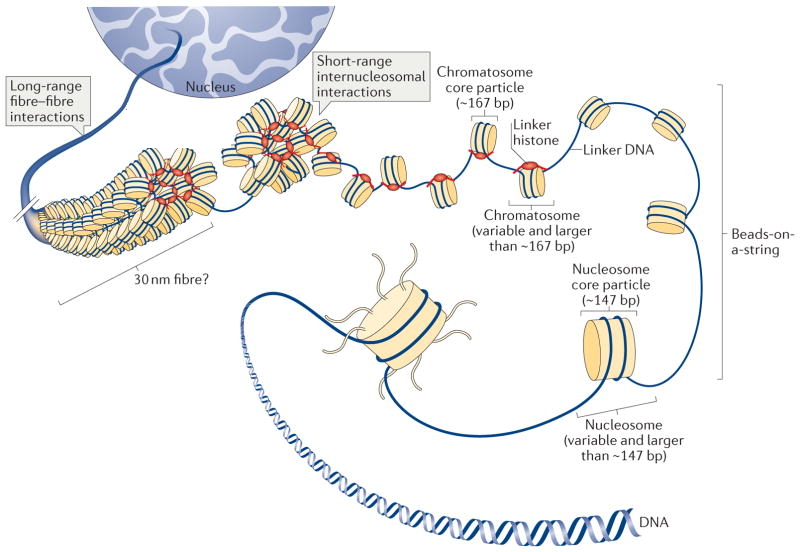
Figure 14.8. Levels of chromatin folding and compaction. Here the “chromatosome core particle” refers to the nucleosome core particle with linker H1 added. (However, all such histone-plus-DNA configurations can still be referred to as “nucleosomes.”) The abbreviation “bp” refers to nucleotide base pairs, so that “167 bp” and “147 bp” refer to the approximate length of DNA wrapped around nucleosomes with and without linker histones, respectively. DNA is ever more fully compacted as the nucleosomes are packed more tightly together. For simplicity, DNA-bound proteins other than histones are not shown. Also, only histone-DNA associations on a single chromatin fiber (chromosome) are depicted here, not associations among different chromosomes.21
“Ribbon” images of the nucleosome core particle, as in Figure 14.9, though highly schematic, are intended to signify certain abstract features of the histone protein structure. The DNA encircling the histones is shown, cartoon-like, in purple.
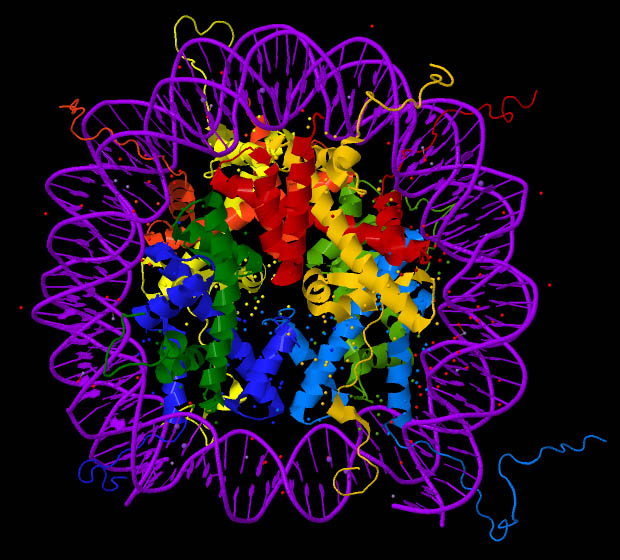
Figure 14.9. A “ribbon” representation of nucleosome structure.22
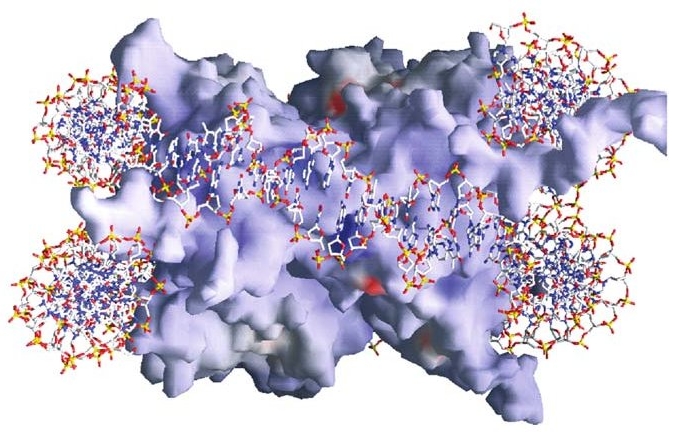
Figure 14.10. Yet a different way to represent the structure of a nucleosome. See main text.23
And yet again, though still with extreme artificiality in terms of the visual image, we have representations such as Figure 14.10, which are generated using data from sophisticated molecular imaging techniques. The red, white, and blue stick figure represents the DNA encircling (about one and two-thirds times) the histone core particle. Red and blue patches on the core particle represent acidic and basic areas, respectively. These, via their effect on the distribution of electrostatic charge over the surface of the histones, have a bearing on many of the functional aspects of the nucleosome discussed below.
Here it is well to remember one of the primary lessons of twentieth-century physics: we are led disastrously astray when we try to imagine atomic- and molecular-level entities as if they were tiny bits of the stuff of our common experience. It would be far better to think of the core particle’s “substance,” “surface,” “contact points,” and “physical interactions” as forms assumed by mutually interpenetrating forces in their intricate and infinitely varied play.
In particular, as geneticist Bryan Turner of the School of Cancer Sciences at the University of Birmingham (UK) reminds us, the nucleosomal core particle “is much more flexible than the crystal structure [which is the basis for images like Figure 14.10] might lead us to believe,” and our current understanding of it “does not lend itself to simplifying generalisations” (Turner 2014). As we will see, the impressive enactments of form and force about the nucleosome are central to any understanding of gene function.
Every “thing” in biology is really an activity, or is caught up in activity, and the extraordinarily dynamic nucleosome is no exception. For example, nucleosomes are the primary feature of chromatin that, as we noted earlier, must be disassembled, or at least “remodeled,” during gene transcription, and then restored to a fully functional state after the transcribing enzyme (RNA polymerase) has passed by.
More generally, the individual histones in a nucleosome can come and go at an almost alarming rate — with an average exchange time of just a few minutes for many nucleosomes. And in some situations the histones exchanged in this way can be different histones — known as “histone variants” — with each variant exerting its own distinct sort of influence on gene expression and chromatin dynamics. Individual histones can even be removed from a core particle altogether, leaving it “incomplete” and now with seriously altered function.
Further: in the course of its life the cell can, and does, reposition huge numbers of nucleosomes along the double helix, bringing to bear upon them a whole galaxy of regulatory interactions. The positioning of nucleosomes — which may be achieved by protein complexes that slide the DNA around the core particle — matters at a highly refined level: a shift by as little as two or three bases (two or three “letters” of the “genetic code”) can make the difference between an expressed or silenced gene (Martinez-Campa et al. 2004). (Individual genes typically contain thousands of bases.)
Still further: not only the exact position of a nucleosome along the double helix, but also the precise rotation of the helix in its embrace of the histones is important. “Rotation” refers to which part of the DNA double helix faces toward a histone surface and which part faces outward. Depending on orientation, the nucleotide bases will be more or less accessible to the various gene-activating and repressing factors that recognize and bind to specific sequences.
This in turn relates to the fact that there are two grooves (the major and minor grooves) running the length of the double helix (Figure 14.11). Proteins that recognize a particular sequence of nucleotide bases typically do so in the major groove, where the sequence is most readily accessible.
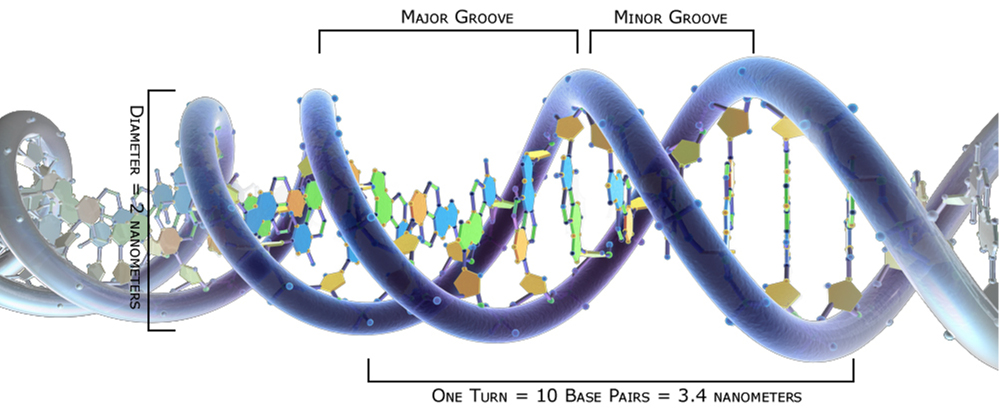
Figure 14.11. A schematic representation of the DNA double helix, showing the major and minor grooves.24
However, many proteins bind to DNA in highly selective ways that can be determined by factors other than the exact DNA sequence. For example, investigations have shown that the minor groove may be compressed so as to enhance the local negative electrostatic potential. Regulatory proteins “read” the compression and the electrostatic potential as cues for binding to the DNA. The “complex minor-groove landscape” (Rohs et al. 2009) is indeed affected by the DNA sequence, but also by associated proteins. Regulatory factors “reading” the landscape can hardly do so according to a strict digital code. By our musical analogy: it’s less a matter of identifying a precise series of notes than of recognizing a melodic and harmonic motif performed by a full orchestra.
You can see, then, why one molecular biologist has referred to the “bewildering array of molecular mechanisms that have evolved to alter the physical properties of nucleosomes” and thereby to play a role in gene regulation (Cosgrove 2012). Also consider this:
Influences such as DNA methylation, posttranslational modifications of the core histone proteins, histone variants, [histone gene] mutations and the level of chromatin compaction may each contribute to a multitude of additional energy states within the chromatin network. All these factors can potentially alter intra- and internucleosomal forces and establish a different or more extended ensemble of nucleosome conformational states, and therefore further fine-tune the functional activities. This is consistent with the notion of a heterogeneous population of nucleosomes within chromatin, all in a dynamic state and able to respond to continuous changes from environmental ques [sic] (Joshi et al. 2012).
But our story of nucleosome-based regulation has so far been radically incomplete.
A tale of tails
We will now look more closely at those parts of the nucleosome where it may be that the most dramatic story unfolds. Refer back to Figure 14.9, representing a nucleosome. The eight histones of the core particle are shown as a ribbon diagram, with the DNA double helix (schematically depicted in purple) wrapped around it somewhat less than two times. You will note a number of thin yellow, red, blue, or green “pig’s tails” extending outward from the core histones. These are the thin, flexible, and mobile histone tails, ten of which are present in the typical core particle. There are hundreds of distinct chemical modifications of these tails (referred to as post-translational modifications), and the countless resulting patterns of modification within any given nucleosome or group of nucleosomes are intimately bound up with the expression of genes. In fact, there is little relating to gene regulation, DNA replication, chromatin structure and dynamics, or the overall functional organization of the nucleus that is not correlated in one way or another with patterns of histone tail modifications.
Learning about these tails, we may be reminded (albeit in a highly fanciful manner) of both the sensory functions of insect antennae and the motor functions of limbs. On the “sensory” side, the tails are receivers of molecular signals coming from all directions in the form of post-translational modifications. The nucleosome provides a context where the integrated significance of these signals can be “read off” (to use the standard phrase) by the gene-regulatory proteins that are sensitive to them. These readers may then “recruit” (again standard usage) various other proteins that either help to restructure chromatin in one way or another, or more directly regulate the expression of genes.
There are in fact many protein “readers” that interact with single modifications, or with groups of them, or with the asymmetrically modified tails of a histone pair, or with a histone modification in proximity to a site of DNA methylation. Every such reader protein acts out of its own world of biochemical genesis, folding, post-translational modification, and conformational plasticity, and together these proteins tell an important part of the story of gene regulation.
Finally, the tails can also act with a kind of brute force as “muscular” effectors. They can, for example — no doubt depending at least in part on their various modifications and protein associations — insinuate themselves into one of the grooves of the double helix, thereby loosening the DNA from the nucleosomal core particle (and making it more available for transcription), or else binding it more tightly. In both cases, one way this is accomplished is by altering the electrical interaction between histone and DNA.
Some of those tails are also thought to establish nucleosome-to-nucleosome contacts, helping to compact a stretch of chromatin. How and whether this is done can make genes either more or less accessible for transcription and various forms of regulation.
Perhaps you can now see why the members of one research team, writing about histone tail modifications, find themselves reflecting upon
the incredibly intricate nature of the chromatin landscape and resultant interactions. The biological consequences of [interactions between histone tail modifications and regulatory proteins] are highly context dependent, relying on the combinatorial readout of the spatially and temporally fluctuating local epigenetic environment and leading to a highly fine-tuned [regulation] of particular genomic sites (Musselman et al. 2012).
A still closer look
We have progressively magnified our field of view by shifting from the overall structure of chromatin, to the nucleosome with its histone core, and then to the individual histone tails. Important principles of gene regulation operate at each different level. Now, magnifying our view one last time, we will home in on a single histone tail modification. The most commonly discussed modifications are the acetylation and methylation of certain lysine amino acids in the tails, but there are many other kinds of modification. Here I will focus on the modification called ubiquitination simply because its gene regulatory roles do not seem quite as extensive (or just are not as well investigated) as those performed by some other tail modifications. This makes their description here a little more manageable.
Monoubiquitination is the “attachment” (a poor word, as I indicated above) of a single ubiquitin chemical group to a lysine amino acid of a protein. In the case of histone tails, this can be done at more than one lysine, but we will look only at the monoubiquitination of lysine 120 on a tail of the histone known as H2B (that is, the lysine at the 120th sequential position along the tail), all of which can be designated H2BK120ub1 (where ‘K’ is the symbol for lysine), but which will be abbreviated here as H2Bub1.
So what is the significance of this modification at a single histone tail location? Here’s one summary:
H2Bub1 takes part in almost every molecular process associated with chromatin biology. H2Bub1 has been shown to regulate transcription initiation and elongation, DNA damage response and repair, DNA replication, nucleosome positioning, RNA processing and export [from the nucleus], chromatin segregation and maintenance of chromatin boundaries. Given the large number of molecular processes regulated by H2Bub1, it is not surprising that H2Bub1 plays a vital role in some of the most fundamental biological processes that occur within multicellular organisms. [Loss of an enzyme responsible for ubiquitination] results in very early embryonic lethality. Furthermore, aberrant H2Bub1 levels can affect cell cycle progression, apoptosis [“programmed cell death”], stem cell differentiation, development, viral infection outcome and "tumorigenesis" (Fuchs and Oren 2014).
(I draw largely on the paper by these authors in the remainder of this section.)
Of course, H2Bub1 does nothing “in general”; results are always specific and context-dependent. For example, blocking this modification in a particular human cell line was found to upregulate some genes, downregulate others, and leave a great many unchanged. Under some circumstances, H2Bub1 is particularly needed for the transcription of relatively long genes. And the modification also plays an important role in histone “crosstalk,” helping to regulate other crucial modifications within the same or on different histones.
A search for “effector” molecules that, singly or cooperatively, associate and interact with the H2Bub1 modification led to the identification of more than ninety proteins, many with known functions in gene regulation consistent with those known to be “effects” of H2Bub1. This points us to what could be a still further extension of our survey, whereby we might analyze one or more of those proteins. We would then have to trace the modifications they undergo, and the larger regulatory world in which they are caught up. But there would be no end of this, since following up any particular line of inquiry in a cell or organism sooner or later leads to everything else.
I have made repeated reference to these ever-widening circles of causal influence. Here I will just momentarily hint at this broader reality in relation to the histone tail modifications called “methylation” (not to be confused with DNA methylation). A methyl group is added to various histone amino acids by enzymes called “methyltransferases,” and is removed by other enzymes called “demethylases.” The mammalian genome is said to encode thirty five histone methyltransferases and twenty three demethylases. This is where the complications enter.
In an article entitled “Controlling the Controllers,” the authors discuss how these methylating and demethylating enzymes are themselves modified and regulated by the addition of phosphoryl groups, with “diverse effect” on enzyme function. Further, the phosphorylation of the enzymes is in turn “regulated by upstream signalling pathways.” And, still further, “different histone methyltransferase and demethylase enzyme families are connected to upstream signalling pathways in different ways” (Separovich 2020). And so the circles widen. But now we must return to our narrower focus.
It remains to mention only that, with ubiquitination as with so many other molecular biological investigations, researchers are vexed by an imagined “need to establish causality more unequivocally” (Fuchs and Oren 2014) — a need that never seems fully satisfied as our understanding grows. This search for unambiguous causes is a fruitless one (Chapter 9, “A Mess of Causes”) because the kinds of causes being looked for don’t exist in organisms.
As for the relations that do exist in organisms, just reflect for a moment. Think, for example, of the transcription network vaguely depicted in Figure 14.1. Then think of the networks of hundreds of mutually regulating mRNAs and microRNAs also discussed above. And now consider the virtually infinite combinations of histone tail modifications and their endlessly elaborated meanings and pervasive “crosstalk.” Many other domains of gene regulation have been alluded to in preceding sections, and untold others could have been mentioned. And now ask yourself what all this must mean. There seem only two possibilities: complete bedlam and chaos of causes working at cross-purposes, or else the play of a coherent, unified, and encompassing wisdom whose all-embracing effectiveness and power of coordination we can hardly yet even begin to conceive.
Movement and rhythm
Few if any details of nucleosome structure and dynamics are fixed and constant. Nothing illustrates this more vividly than the fact of DNA breathing on the nucleosome surface. This refers to the partial and rhythmical unwrapping and re-wrapping of the double helix, especially near the points of entry and exit on the nucleosome. This provides what are presumably well-gauged, fractional-second opportunities for gene-regulating proteins to bind to their target DNA sequences during the periods of relaxation:
Some transcription factors (TFs) only recognize nucleosomal DNA when nucleosome “breathing” occurs, that is when the DNA is partially and temporarily unwrapped from the nucleosome surface … histone post-translational modifications facilitate DNA breathing. TF binding facilitates further nucleosome unwrapping by promoting the binding of additional TFs, and/or in coordination with chromatin remodelers. Some TFs can bind their cognate motifs on fully compacted nucleosomal DNA and initiate ATP-independent DNA unwrapping or even histone eviction. However, outcomes in which TF binding stabilizes nucleosomes are also possible (Makowski, Gaullier and Luger 2020).
This breathing also relates to the transcriptional pausing by RNA polymerase (discussed above). The polymerase appears able to take advantage of the breathing in order to move, step by step and with significant pauses, along the genes it is transcribing. In this way the characteristics of nucleosomes — how the DNA breathes, and whether it is firmly or loosely anchored to a histone at any particular moment and place — can affect the timing and frequency of pauses. And, as we saw earlier, the rhythm of pauses and movements then affects the splicing and folding of the RNA being synthesized, which in turn bear on how the RNA can be regulated as well as the structure and function of the protein molecule produced from the RNA. A proper “music” is required for the overall performance to be successful. So it appears that the references to “choreography” and “dance” one sometimes encounters in the literature may be more than mere poetic niceties.
With a different sort of rhythm nucleosomes will sometimes move — or be moved (as I have remarked before, the distinction between “actor” and “acted upon” is forever obscured in the living cell) — rhythmically back and forth along the DNA, shifting between alternative positions in order to enable multiple transcriptional passes over a gene by RNA polymerase.
Stem cells exhibit what some have called “histone modification pulsing,” which results in the continual application and removal of both gene-repressive and gene-activating modifications of nucleosomes. In this way a delicate balance is maintained around genes involved in development and cell differentiation. The genes are kept, so to speak, in a finely poised state of “dynamic and balanced readiness,” so that when the decision to specialize is finally taken, the repressive modifications can be quickly lifted, leading to rapid gene expression (Gan et al. 2007).
This state of suspended readiness in stem cells also seems to be served by a rhythmical (10 – 100 cycles per second), back-and-forth spatial movement, or vibration, of chromatin within the cell nucleus. Associated with “hyperdynamic binding of structural proteins” mediated by nucleosomes, this vibration is thought to help maintain the largely open chromatin state characteristic of stem cells. The movement depends on the metabolic state of the cell and is progressively dampened as the stem cell differentiates into a specialized cell with substantial portions of its chromatin in a condensed state (Hinde 2012).
Box 14.1
From Static Mechanism to Dynamic Regulator
In an article entitled “Understanding Nucleosome Dynamics and Their Links to Gene Expression and DNA Replication,” Pennsylvania State University molecular biologists William Lai and Franklin Pugh concluded their review of nucleosomes this way:
“Originally viewed as a rather static mechanism of chromatin packaging, the nucleosome core complex is now well recognized as one of the key regulatory components of the genome. We also now see that instead of static protein complexes, nucleosomes are in fact exceptionally dynamic and that their positioning and composition are crucial for genome regulation. As such, the study of nucleosome dynamics is essentially the study of genome regulation. The complex interaction between nucleosome occupancy and positioning allows the cell to properly regulate accessibility of various proteins and their complexes to DNA and thus to regulate gene expression programmes. A variety of regulatory cofactors such as chromatin remodellers, chaperones and general regulatory factors operates both independently and synergistically to maintain the precise organization and composition of nucleosome arrays at specific genomic loci. This dynamic environment probably exists so that the genome may respond and adapt quickly to both external stimuli as well as be able to quickly recover from chromatin-disruptive activities such as transcription and replication” (Lai 2017).
With reference to that last sentence, it needs adding that what “responds and adapts quickly” to external and internal stimuli is not really the rather passive genome so much as the entire, all-encompassing regulatory environment, of which the nucleosome is a neat picture and summary.
But quite apart from stem cells, it is increasingly appreciated that nucleosomes play a key role in holding a balance between the active and repressed states of genes in many cell types. As the focus of a highly dynamic conversation involving histone variants, histone tail modifications, and innumerable chromatin-associating proteins, decisively placed nucleosomes can (as biologist Bradley Cairns writes) maintain genes “poised in the repressed state,” and “it is the precise nature of the poised state that sets the requirements for the transition to the active state.” Among other aspects of the dynamism, there is continual turnover of the nucleosomes themselves — and of their separate components — a turnover that allows transcription factors to gain access to DNA sequences “at a tuned rate” (Cairns 2009).
It is perhaps worth mentioning here that in certain bacteria a 24-hour (circadian) rhythm correlates with the changing state of DNA supercoiling — that is, with a tighter or looser twisting of the double helix. It appears that something similar may be going on in higher animals, where DNA supercoiling is so closely “wrapped up” with nucleosomes. In these organisms one of the factors involved in the extremely complex processes by which genes are regulated in a circadian fashion is the rhythmic application of histone modifications to selected nucleosomes (Woelfle et al. 2007), presumably with direct implications for chromatin structure and DNA supercoiling.
The nucleosome, we can fairly say, is a ceaselessly transforming matrix and organizational hub whose structure and pattern of activity is never exactly duplicated anywhere in the genome. It is where the infinitely ramified interface between the larger cell and its DNA comes to its most focal expression. And that expression turns out to be livingly nuanced activity, dynamic beyond what anyone imagined during the age of the double helix as the one-dimensional “secret of life.”
And so, seemingly in the grip of the encircling DNA with its relatively fixed and stable structure, yet responsive to the ceaselessly varying flows of life around it, the nucleosome holds a muscular and intelligent balance between gene and context — a task requiring flexibility and a play of appropriate rhythm (Box 14.1).
Such, then, is the intimate, intricate, well-timed choreography through which our genes come to their proper expression. And the plastic, shape-shifting nucleosome in the middle of it all provides an excellent vantage point from which to view the overall drama of form and movement.
A story mostly untold
We have, in our review, only sparsely sampled the overwhelming number of causal factors participating in gene expression. The topics not touched upon here — the unmentioned domains of regulatory, or epigenetic, activity affecting what the cell makes of its genes — would extend the presentation vastly beyond the topics I have briefly alluded to here.
There is, for example, the recently intensifying exploration of the importance of modifications, not only on the histone tails, but also on the histone cores. These also are proving relevant to gene expression, and in complex ways, both direct and roundabout.
We could also have talked about the entire universe of regulation governing the translation of mRNA molecules into protein after they have been exported from the cell nucleus into the cytoplasm. The task is accomplished by complexes of protein and RNA known as “ribosomes.” The diverse factors the cell gathers together for translation rival those we see in gene transcription.
And once a protein is generated, there is the problem of its folding (and re-folding), often with the help of “chaperone” proteins. Many proteins can potentially fold in an almost unlimited number of ways, yet achieving the “right” folds is crucial for protein function. This folding of a protein can begin already as it is being translated from RNA. Moreover, the folding outcome may be affected by the innumerable factors playing into the activity of translation. We do not often find just one thing at a time being accomplished by any biological process. (Something similar is true of RNAs. We have seen that both alternative splicing and folding of an RNA can occur — with major functional implications — during its transcription from DNA.)
Then, still further downstream from gene transcription, there are the various post-translational modifications (PTMs) that may be applied, removed, and re-applied to any gene-regulatory protein (transcription factors, co-activators, co-repressors, chromatin remodelers, and so on), just as we saw with the histone proteins belonging to nucleosomes. These again shape the molecule’s function, often in a dynamic, ever-shifting way as the modifications come and go. Together, the many thousands of proteins subject to PTMs, and the diverse effects of these modifications, make for a vast regulatory landscape almost impossible to encompass in thought. The resulting regulatory activity is always context-dependent, relating to larger, governing purposes rather than being the mere effect of a local physical necessity.
We could also talk about what is, in one sense, the most fundamental biological activity of all — metabolism. After all, every performance of our body derives in one way or another from the food we eat. Metabolites and the organization of metabolic processes play critical roles in many aspects of gene expression related to everything from circadian rhythms to cancer.
Or we could talk about how some RNAs, especially non-protein-coding RNAs, form a “scaffolding” that gives structure to the cell nucleus and therefore plays a fundamental role in just about all nuclear functions. Except that words such as “scaffolding” and “structure” can be very misleading, as two researchers point out in a paper entitled “Role of Nuclear RNA in Regulating Chromatin Structure and Transcription.” We should expect, they write, that “any nuclear structure that is assembled employing RNA cannot be static but [must be] constantly recycling degraded RNA with newly synthesised ones.” So “the original concept of a static nuclear matrix must be re-evaluated in terms of a dynamic scaffold” (Michieletto and Gilbert 2019).
Perhaps the most intense and significant, newer field of research bearing on gene regulation in recent years relates to phase transitions in the cell, and especially in the nucleus. (See Chapter 5, “Our Bodies Are Formed Streams.”) Like ice crystals forming and dissolving in water held near the freezing point, or like oil droplets in some other liquid (or like water droplets in oil), complex combinations of proteins, RNAs, and other molecules can form separated-out liquid or semi-solid aggregates (droplets) within the cellular plasm. The dynamic functional role of these aggregates in bringing molecular communities together at the right place, in the right amounts, and at the right time is now a prime topic relating to just about everything discussed in this chapter. The new understanding we are gaining in this field makes a mechanistic or deterministic interpretation of cellular physiology even less tenable than it already was.
And if any new topic of research ranks second to phase transitions in importance, it surely must be the one focusing on the role of the microbiome. The total DNA sequence of all the microorganisms in our bodies exceeds that of the trillions of cells in our bodies. The processess rooted in this “foreign” DNA can affect our biology, much as can the processes stemming from our own DNA. And the effects extend to regulation of our genes.
But surely it is time for us to stop. Anyone desiring a glimpse of the wider range of topics relating to gene expression might wish to scan the expanded outline of topics near the beginning of the article, “How the Organism Decides What to Make of Its Genes (Talbott 2021).
Concluding thoughts
A decisive problem for the classical view of DNA is that a human cell employs its 20,000 or so genes to generate an estimated 250,000 to 1 million distinct proteins (Klerk and ’t Hoen 2015). The activities shaping these abundant outcomes are not strictly determined by DNA. Rather, they arise from all corners of the cell and larger organism, just as the outcomes themselves — all those distinct proteins — are ushered to their proper places in every cell of every tiniest niche throughout the whole. We are always watching integral and unified performances. The idea that genes are originating causes that make everything else happen is grotesquely wrong-headed.
Mina Bissell, a researcher who has received many recognitions, has, along with her co-author, put the matter this way: “The sequence of our genes are [sic] like the keys on the piano; it is the context that makes the music” (Bissell and Hines 2011). We might add that the raw DNA sequence does not even contain all the keys; let’s say: just the white keys. The flats and sharps, without which the music would lose its savor, are provided by DNA methylation, RNA editing, and so much more.
And Shelley Berger, the Daniel S. Och professor of cell and developmental biology at the University of Pennsylvania School of Medicine’s Wistar Institute — after noting that a single histone tail modification “recruits numerous proteins whose regulatory functions are not only activating but also repressing,” and that “many of these marks have several, seemingly conflicting roles” — summarized the situation this way:
Although [histone] modifications were initially thought to be a simple code, a more likely model is of a sophisticated, nuanced chromatin “language” in which different combinations of basic building blocks yield dynamic functional outcomes (Berger 2007).
What she says about histone tail modifications could just as well be said, as we have seen, about the entire universe of gene regulation. We are looking at a meaningful, qualitative, and thoughtful language through which living narratives are constructed. In slightly different terms, Berger envisions histone modifications as participating in “an intricate ‘dance’ of associations.”
In the plastic organism, what goes on at the local level is always shaped and guided by a larger, coherent context — a context that surely has meaning, but (as in natural languages) never an absolutely fixed grammar or logic. And, in fact, while overwhelming evidence for a meaningful, gene-regulatory conversation involving histone modifications has emerged, there is little to suggest a rigid code — this despite the strong urge in molecular biologists to find one.
The overall picture of gene expression is one of unsurveyable complexity in the service of remarkably effective living processes. What all the foregoing shows is that the whole cell and the whole organism are forever carrying out narrative tasks. We have no explanatory coherence so long as we are following individual chains of molecular causation. The mutually interpenetrating lines of influence converging upon and issuing from our DNA reveal their full meaning only when we consider what needs and interests are reflected in the overall, coordinated pattern of causes — what the organism is doing and why.
Gene Expression: A Long and Winding Journey
If you feel exhausted at this point, I will understand. So do I. Any effort to fully take hold of life, at any scale of observation and activity, can prove exhausting. The way in which gene expression arises from, or is disciplined by, or is made to serve, all aspects of an organism’s life may be tiring to explore, even in the sorely incomplete manner of the foregoing. But taking note of the basic fact of the matter is well worthwhile. I am not at all tempted to try to summarize anew here the ground we have covered. But I will extract two statements from the text above suggesting one way to view the significance of everything we have looked at:
(1) Given the play of infinite, interwoven influences at the molecular level, where non-mechanical fluidity rules and the number of actors relevant to just about any function of the cell or organism is unlimited, there seem only two possibilities: complete bedlam and chaos of causes working at cross-purposes, or else the play of a coherent, unified, and encompassing wisdom whose all-embracing effectiveness and power of coordination we can hardly yet even begin to conceive.(2) In the plastic organism, what goes on at the local level is always shaped and guided by a larger, coherent context — a context that surely has meaning, but (as in natural languages) never an absolutely fixed and determining grammar or logic.
These conclusions could hardly be more upsetting for a molecular biology centered on theoretical notions of code, informational logic, and discrete causes. We not only need a tracing of physical and chemical lawfulness, but also an understanding of the meaning, end (telos), and purposiveness of things — a hard pill to swallow for the conventionally trained biologist. But it’s not as if much imagination is required in order to see which way the current is pulling us in today’s deep-diving explorations of molecular biology.
We had an introduction to epigenetics (as genetics seen in context) in Chapter 7. That, together with this current chapter, as well as much else in the first half of the book will need to be kept in mind as we pass on to the discussion of evolution in the second half of the book. We will see that the main point of the older, outmoded concept of gene expression was to eliminate the life of the organism from evolutionary theorizing. If you remember what you have read here, you will have much less difficulty thinking about how organisms themselves — collectively organized in a species or population — might be the real drivers of evolution, much as the cells and microbiome, collectively in each of us, are so organized as to give adaptive expression to the life of the individual.
Notes
1. In Chapter 8 (“The Mystery of an Unexpected Coherence”) we looked at how proteins can rescue completely shattered DNA.
2. The “promiscuity” of binding — that is, binding in the absence of definitive binding sequences — is a problem relating to protein-nucleotide interactions in general. For example, 55 percent of RNA-binding proteins “do not contain any known RNA-binding domain at all” (Editors of Nature Structural & Molecular Biology 2021).
3. Figure 14.1 credit: from Wei, He, Zhang et al. (2021), CC BY-NC-SA 4.0.
4. I will not discuss the RNA portion of chromatin here. But its functions, which researchers are now struggling to unravel, look as though they may rival the diverse functions of the protein portion.
5. No contemporary biologist has a sound basis for assuming “necessary contextualization and direction,” because the idea of wise direction is foreign to the current presuppositions of biology. But every biologist, in talking about specific molecular processes, nevertheless does make the assumption — and makes it for the simple reason that there is no alternative. We either assume the wisely guided context or our immediate work becomes meaningless. It loses its whole point, which is to explain how one or another process contributes to a function or task — that is, to an effectively directed, purposive activity (Chapter 2, “The Organism’s Story”). So biologists are forever implicitly placing themselves within a theoretical framework that, from their own standpoint, is indefensible.
6. By “modest-sized” I mean: about 2000 nucleotide bases in length.
7. Figure 14.2 credit: From Kazantseva and Palm 2014, CC BY-SA 3.0.
8. Figure 14.3 credit: From Tóth-Petróczy et al. 2008, CC BY-SA 4.0.
The article from which the figure was taken concerns the propensity of Mediator proteins to contain “intrinsically disordered” regions. The authors conclude that “conserved intrinsically disordered regions contribute to the gene-specific regulatory function of the Mediator. Intrinsically disordered regions with weak sequence restraints can provide an evolutionarily economic solution for the Mediator to handle a steadily increasing amount of complex regulatory signals.”
9. Here is one paragraph from a paper on the Mediator complex:
The Mediator is an evolutionarily conserved, multiprotein complex that is a key regulator of protein-coding genes. In metazoan cells, multiple pathways that are responsible for homeostasis, cell growth and differentiation converge on the Mediator through transcriptional activators and repressors that target one or more of the almost 30 subunits of this complex. Besides interacting directly with RNA polymerase II, Mediator has multiple functions and can interact with and coordinate the action of numerous other co-activators and co-repressors, including those acting at the level of chromatin. These interactions ultimately allow the Mediator to deliver outputs that range from maximal activation of genes to modulation of basal transcription to long-term epigenetic silencing (Malik and Roeder 2010).
Mediator also has tissue-specific aspects:
Adding yet another degree of complexity, members of the same transcription factor family can target different Mediator subunits to activate transcription of the same gene, through the same promoter elements, in different cell types (Conaway and Conaway 2011).
10. Figure 14.4 credit: From Quevedo et al. (2019), CC BY 4.0.
11. Figure 14.5 credit: RCSB Protein Data Bank (http://www.rcsb.org), courtesy of David S. Goodsell.
12. The Wikipedia article, “Tata-binding protein” (accessed on April 1, 2019), offers a succinct description of part of this interaction: “When TBP binds to a [particular sequence] within the DNA, it distorts the DNA by inserting amino acid side-chains between base pairs, partially unwinding the helix, and doubly kinking it. The distortion is accomplished through a great amount of surface contact between the protein and DNA. TBP binds with the negatively charged phosphates in the DNA backbone through positively charged lysine and arginine amino acid residues. The sharp bend in the DNA is produced through projection of four bulky phenylalanine residues into the minor groove. As the DNA bends, its contact with TBP increases, thus enhancing the DNA-protein interaction.”
13. There are actually three RNA polymerase enzymes in humans: RNA polymerase I, II, and III. I will be speaking of RNA polymerase II, which transcribes the great majority of our genes. Also, “RNA” in the following descriptions will refer either to messenger RNA (mRNA), which can be translated into protein, or else to RNA more generally. References to specific non-protein-coding RNAs such as microRNAs (miRNAs) will be flagged as such.
14. Just about any functional significance of an RNA — from what protein it produces, to its stability and cellular localization, to the various roles of its three-dimensional structure — can be affected by this editing. One kind of editing (known as A-to-I editing) “is extremely abundant in primates: over a hundred million editing sites exist in [RNAs derived from] their genomes” (Levanon and Eisenberg 2014). However, biologists have only begun to explore the functional significance of most of this editing, and there remains among the majority of researchers today a tendency to dismiss as “random noise” whatever their current methods and concepts cannot presently illuminate.
15. Frye 2018. Regarding one of these modifications, known as mRNA adenosine methylation (m6A), Timothy Nilsen, a molecular biologist at Case Western Reserve University in Cleveland, has written:
A series of papers have appeared in rapid succession, together providing a wealth of unequivocal evidence for m6A function. But these findings still have not led to a coherent picture of the number and variety of functions of the m6A modification (Nilsen 2014).
In the years since he wrote that, the picture has, bit by bit, been filled in, and continues to be filled in. But there is a long way to go.
16. The ceRNA network we’re discussing is extremely simple. The authors of the paper presenting it refer to a study of brain cancer (glioblastoma) where “the analysis was significantly extended beyond the binary ceRNA associations described in most other studies,” and “the PTEN ceRNA interactions were found to be part of a post-transcriptional regulatory layer comprising more than 248,000 microRNA-mediated interactions.”
17. Of course, anything can be analyzed in one way or another if we narrow our vision sufficiently and disregard, for example, the purposive (telos-realizing) aspects of what is going on. The question is whether analyzing living activity by breaking it into physically explicable part-processes yields an explanation or understanding of its telos-realizing character. Throughout this book I have been pointing out the incommensurability between a strictly physical analysis of biological phenomena and the recognizable meaning of those phenomena.
18. Figure 14.6 credit: Courtesy of Donald Olins.
19. An example of the functioning of linker histones: “Our results establish H1 as a critical regulator of gene silencing through localized control of chromatin compaction, 3D genome organization and the epigenetic landscape” (Willcockson et al. 2020).
The functions of the linker histone are also indicated by the fact that “mutations in H1 drive malignant transformation primarily through three-dimensional genome reorganization, which leads to epigenetic reprogramming and derepression of developmentally silenced genes” (Yusufova et al. 2020). And then there is this: “The biochemical functions of H1 in the regulation of nuclear DNA metabolism should not be limited to a single, one-size-fits-all DNA compaction paradigm. Rather, H1 appears to be an active biochemical player in chromatin and a potent effector of multiple aspects of chromosome structure and chromatin functions” (Fyodorov 2018).
20. Figure 14.7 credit: Darekk2 (https://commons.wikimedia.org/wiki/File:Nucleosome_organization.png), CC BY-SA 3.0.
21. Figure 14.8 credit: From Fyodorov et al. 2018.
22. Figure 14.9 credit: Darekk2 (https://commons.wikimedia.org/wiki/File:Nucleosome_core_particle_1EQZ_v.5.jpg) based on data from the Protein Data Bank, CC BY-SA 3.0.
23. Figure 14.10 credit: From Luger 2006.
24. Figure 14.11 credit: Zygote Media Group (https://commons.wikimedia.org/wiki/File:3DScience_DNA_structure_labeled_a.jpg), CC BY 2.5.
Sources
Berger, Shelley L. (2007). The Complex Language of Chromatin Regulation during Transcription, Nature vol. 447 (May 24), pp. 407-12. doi:10.1038/nature05915
Bissell, Mina and William C. Hines (2011). Why Don’t We Get More Cancer? A Proposed Role of the Microenvironment in Restraining Cancer Progression, Nature Medicine vol. 17, no. 3 (March), pp. 320-29. doi:10.1038/nm.2328
Bobola, Nicoletta and Samir Merabet (2017). Homeodomain Proteins in Action: Similar DNA Binding Preferences, Highly Variable Connectivity, Current Opinion in Genetics and Development vol. 43 (April), pp. 1-8. doi:10.1016/j.gde.2016.09.008
Brown, Valerie (2008). Environment Becomes Heredity, Pacific Standard (July 14). https://psmag.com/nature-and-technology/environment-becomes-heredity-4425
Cairns, Bradley R. (2009). The Logic of Chromatin Architecture and Remodelling at Promoters, Nature vol. 461 (September 10), pp. 193-98. doi:10.1038/nature08450
Chaires, Jonathan B. (2008). Allostery: DNA Does It Too, ACS Chemical Biology vol. 3, no. 4 (April 18), pp. 207-9. doi:10.1021/cb800070s
Conaway, Ronald C. and Joan Weliky Conaway (2011). Function and Regulation of the Mediator Complex, Current Opinion in Genetics and Development vol. 21, pp. 225-30. doi:10.1016/j.gde.2011.01.013
Cosgrove, Michael (2012). Writers and Readers: Deconvoluting the Harmonic Complexity of the Histone Code, Nature Structural and Molecular Biology vol. 19, no. 8 (August), pp. 739-40. doi:10.1038/nsmb.2350
Editors of Nature Molecular & Structural Biology (2021). The Nucleotides That Bind, Nature Structural & Molecular Biology vol. 28, no. 1 (January). doi:10.1038/s41594-020-00552-8
Frye, Michaela, Bryan T. Harada, Mikaela Behm and Chuan He (2018). RNA Modifications Modulate Gene Expression during Development, Science vol. 361, no. 6409 (September 28), pp. 1346-49. doi:10.1126/science.aau1646
Fuchs, Gilad and Moshe Oren (2014). Writing and Reading H2B Monoubiquitylation, Biochimica et Biophysica Acta vol. 1839, pp. 694–701. doi:10.1016/j.bbagrm.2014.01.002.
Fyodorov, Dmitry V., Bing-Rui Zhou, Arthur I. Skoultchi and Yawen Bai (2018). Emerging Roles of Linker Histones in Regulating Chromatin Structure and Function, Nature Reviews Molecular Cell Biology vol. 19, no. 3 (March), pp. 192-206. doi:10.1038/nrm.2017.94
Gan, Qiong, Tadashi Yoshida, Oliver G. McDonald et al. (2007). Concise Review: Epigenetic Mechanisms Contribute to Pluripotency and Cell Lineage Determination of Embryonic Stem Cells, Stem Cells vol. 25, no. 1, pp. 2-9. doi:10.1634/stemcells.2006-0383
Hill, Meredith and Nham Tran (2021). Global miRNA to miRNA Interactions: Impacts for miR-21, Trends in Cell Biology vol. 31, no. 1, pp. 3-5. doi:10.1016/j.tcb.2020.10.005
Hinde, Elizabeth, Francesco Cardarelli, Aaron Chen et al. (2012). Tracking the Mechanical Dynamics of Human Embryonic Stem Cell Chromatin, Epigenetics and Chromatin vol. 5, no. 20. doi:10.1186/1756-8935-5-20
Joshi, Sachindra R., Yaw C. Sarpong, Ronald C. Peterson and William M. Scovell (2012). Nucleosome Dynamics: HMGB1 Relaxes Canonical Nucleosome Structure to Facilitate Estrogen Receptor Binding, Nucleic Acids Research vol. 2012, pp. 1-11. doi:10.1093/nar/gks815
Kazantseva, Jekaterina and Kaia Palm (2014). Diversity in TAF Proteomics: Consequences for Cellular Differentiation and Migration, International Journal of Molecular Sciences vol. 15, no. 9 (September 19), pp. 16680-97. doi:10.3390/ijms150916680
Klerk, Eleonora de and Peter A. C. ’t Hoen (2015). Alternative mRNA Transcription, Processing, and Translation: Insights from RNA Sequencing, Trends in Genetics vol. 31, no. 3 (March), pp. 128-39. doi:10.1016/j.tig.2015.01.001
Kristjánsdóttir, Katla, Elizabeth A. Fogarty and Andrew Grimson (2015). Systematic Analysis of the Hmga2 3’ UTR Identifies Many Independent Regulatory Sequences and a Novel Interaction Between Distal Sites, RNA vol. 21, no. 7 (July), pp. 1346-60. https://rnajournal.cshlp.org/cgi/doi/10.1261/rna.051177.115
Kwak, Hojoong and John T. Lis (2013). Control of Transcriptional Elongation, Annual Review of Genetics vol. 47, pp. 483-508. doi:10.1146/annurev-genet-110711-155440
Lai, William K. M. and B. Franklin Pugh (2017). Understanding Nucleosome Dynamics and Their Links to Gene Expression and DNA Replication, Nature Reviews Molecular Cell Biology vol. 18, pp. 548-62. doi.org/10.1038/nrm.2017.47
Levanon, Erez Y. and Eli Eisenberg (2014). Does RNA Editing Compensate for Alu Invasion of the Primate Genome?, Bioessays vol. 37, pp. 175–81. https://doi.wiley.com/10.1002/bies.201400163
Luger, Karolin (2006). Dynamic Nucleosomes, Chromosome Research vol. 14, pp. 5-16. doi:10.1007/s10577-005-1026-1
Makowski, Matthew M., Guillaume G. Aullier and Karolin Luger (2020). Picking a Nucleosome Lock: Sequence- and Structure-Specific Recognition of the Nucleosome, Journal of Biosciences vol. 45, article 13. doi:10.1007/s12038-019-9970-7
Malik, Sohail and Robert G. Roeder (2010). The Metazoan Mediator Co-Activator Complex as an Integrative Hub for Transcriptional Regulation, Nature Reviews Genetics vol. 11 (November), pp. 761-72. doi:10.1038/nrg2901
Martinez-Campa, Carlos, Panagiotis Politis, Jean-Luc Moreau et al. (2004). Precise Nucleosome Positioning and the TATA Box Dictate Requirements for the Histone H4 Tail and the Bromodomain Factor Bdf1, Molecular Cell vol. 15 (July 2), pp. 69-81. doi:10.1016/j.molcel.2004.05.022
Michieletto, Davide and Nick Gilbert (2019). Role of Nuclear RNA in Regulating Chromatin Structure and Transcription, Current Opinion in Cell Biology vol. 58, pp. 120-25. doi:10.1016/j.ceb.2019.03.007
Miller, Courtney A. and J. David Sweatt (2007). Covalent Modification of DNA Regulates Memory Formation, Neuron vol. 53 (March 15), pp. 857-69. doi:10.1016/j.neuron.2007.02.022
Moretti, Rocco, Leslie J. Donato, Mary L. Brezinski et al. (2008). Targeted Chemical Wedges Reveal the Role of Allosteric DNA Modulation in Protein-DNA Assembly, ACS Chemical Biology vol. 3, no. 4, pp. 220-29. doi:10.1021/cb700258r
Musselman, Catherine A., Marie-Eve Lalonde, Jacques Côte and Tatiana G. Kutaleladze (2012). Perceiving the Epigenetic Landscape Through Histone Readers, Nature Structural and Molecular Biology vol. 19, no. 12 (December), pp. 1218-27. doi:10.1038/nsmb.2436
Mustoe, Anthony M., Charles L. Brooks and Hashim M. Al-Hashimi (2014). Hierarchy of RNA Functional Dynamics, Annual Review of Biochemistry vol. 83, pp. 441–66. doi:10.1146/annurev-biochem-060713-035524
Nilsen, Timothy W. (2014). Internal mRNA Methylation Finally Finds Functions, Science vol. 343 (March 14), pp. 1207-8. doi:10.1126/science.1249340
Quevedo, Marti, Lize Meert, Mike R. Dekker, et al. (2019). Mediator Complex Interaction Partners Organize the Transcriptional Network That Defines Neural Stem Cells, Nature Communications vol. 10, no. 2669. doi:10.1038/s41467-019-10502-8
Rohs, Remo, Sean M. West, Alona Sosinsky et al. (2009). The Role of DNA Shape in Protein-DNA Recognition, Nature vol. 461 (October 29), pp. 1248-53. doi:10.1038/nature08473
Separovich, Ryan J., Chi Nam Ignatius Pang, and Marc R. Wilkins (2020). Controlling the Controllers: Regulation of Histone Methylation by Phosphosignalling, Trends in Biochemical Sciences. https://doi.org/10.1016/j.tibs.2020.08.004
Severin, Philip M. D., Xueqing Zou, Hermann E. Gaub and Klaus Schulten (2011). Cytosine Methylation Alters DNA Mechanical Properties, Nucleic Acids Research vol. 39, no. 20, pp. 8740-51. doi:10.1093/nar/gkr578
Slattery, Matthew, Tianyin Zhou, Lin Yang et al. (2014). Absence of a Simple Code: How Transcription Factors Read the Genome, Trends in Biochemical Sciences vol. 39, no. 9 (September), pp. 381-99. doi:10.1016/j.tibs.2014.07.002
Talbott, Stephen L. (2021). How the Organism Decides What to Make of Its Genes. https://bwo.life/org/support/genereg.htm
Tay, Yvonne, John Rinn and Pier Paolo Pandolfi (2014). The Multilayered Complexity of ceRNA Crosstalk and Competition, Nature vol. 505 (January 16), pp. 344-52. doi:10.1038/nature12986
Tóth-Petróczy, Ágnes, Christopher J. Oldfield, István Simon et al. (2008). Malleable Machines in Transcription Regulation: The Mediator Complex, PLOS Computational Biology (December 19). doi.org/10.1371/journal.pcbi.1000243
Turner, Bryan M. (2014). Nucleosome Signalling: An Evolving Concept, Biochimica et Biophysica Acta vol. 1839, pp. 623-26. doi:10.1016/j.bbagrm.2014.01.001
Wei, Li, Fei He, Wen Zhang, Wenhua Chen, and Bo Yu (2021). Analysis of Master Transcription Factors Related to Parkinson’s Disease Through the Gene Transcription Regulatory Network, Archives of Medical Science vol. 17, no. 5.
Wei, Jiufeng, Guodong Li, Shuwei Dang et al. (2016). Discovery and Validation of Hypermethylated Markers for Colorectal Cancer, Disease Markers vol. 2016. doi:10.1155/2016/2192853
Willcockson, M. A., S. E. Healton, C. N. Weiss et al. (2020). "H1 Histones Control the Epigenetic Landscape by Local Chromatin Compaction, Nature (December 9). https://doi.org/10.1038/s41586-020-3032-z
Woelfle, Mark A., Yao Xu, Ximing Qin et al. (2007). Circadian Rhythms of Superhelical Status of DNA in Cyanobacteria, PNAS vol. 104, no. 47, pp. 18819-24. doi:10.1073/pnas.0706069104
Yusufova, Nevin, Andreas Kloetgen, Matt Teater et al. (2020). Histone H1 Loss Drives Lymphoma by Disrupting 3D Chromatin Architecture, Nature (December 9). https://doi.org/10.1038/s41586-020-3017-y
This document: https://bwo.life/bk/epigene2.htm
Steve Talbott :: How Our Genes Come to Expression
