Biology Worthy of Life
An experiment in revivifying biology
Are “Disordered” Proteins Really Disordered?
Stephen L. TalbottIf there’s one lesson current work in molecular biology is drilling into us, it is that organic form never exhibits the mechanical qualities that have been the rather odd starting point for biological thinking over the past several decades. This is true whether we’re talking about the form of things (including molecules) or of processes. I’ve previously written briefly about how the “same” proteins, in different environments, can be viewed as completely different molecules, with different properties. Likewise, I’ve written about the “horror graphs” resulting from the complex cross-talk of signaling pathways and also about the fluidity of living processes in general.
Quite apart from the malleability and shifting conformation of proteins that are considered more or less well-structured, biologists are now paying attention to the substantial portion of all the proteins in our bodies that are “intrinsically disordered”. But that standard phrase could hardly be more misleading, for there is clearly method in the disorder. In fact, these proteins are being found crucial to processes for which dynamic form is key. That, of course, could mean virtually all processes in the organism, but the current focus is on the role intrinsically disordered proteins play as Proteus-like hub elements in protein interaction networks, and as critical factors in molecular signaling.
Many intrinsically disordered proteins are promiscuous binders that interact with multiple partners and frequently function as molecular hubs in protein interaction networks.
That’s an opening remark by Ferreon et al.* in a paper showing the remarkable role of one particular protein known as E1A. Derived from a virus, E1A interacts with a variety of molecular partners to alter the epigenetic state of the cell and thereby influence gene expression. In different organisms it can serve either as a carcinogen (mouse) or as a tumor suppressor (humans).
The E1A protein has two distinct regions, either of which may shift between disordered and ordered forms. Depending on such shifts, as well as on the combination of other molecules that bind to the two regions — molecules that may, under different conditions of E1A “disorder”, have either cooperative or antagonistic effects upon the function of E1A — very different things may happen. As one researcher summarizes the matter:
Intrinsically disordered proteins ... can not only regulate the magnitude of a particular signal, but also reverse it, transforming a positive effector into a negative one. This result reinforces an emerging view that such proteins play a crucial part in signalling, and challenges much of the current dogma about the relationship between protein structure and function ... (Hilser 2013*).
It is ironic that, when molecular forms show themselves to be engaged in much more intensive and dynamic forming — performing — than we could expect of static, crystalline structures, they should be called “disordered”. “Eloquent” might be closer to the reality. But the eloquence has easily been silenced — by the straightforward expedient of turning a deaf ear toward it. As another research team has written: intrinsically disordered regions of protein molecules “have inherent dynamic properties that allow them to ‘scan’ for different binding partners depending on the activation state of the cell” — but this only causes trouble for anyone who is trying to establish a molecular structure. The molecule, so to speak, won’t stand still for its photograph.
The simple solution? Remove the troublesome, dynamic part of the molecule so that we can obtain the kind of crystalline picture we like: “The tendency is to simply delete these regions [in order] to conduct structural studies, even though IDRs [intrinsically disordered regions] are extremely important for biological function and regulation” (Taylor et al. 2012*).
The urge to see nature as consisting of tiny machines — and the surprise at finding things otherwise — seems particularly pronounced among biologists. Perhaps it is because they worry that we may notice how vital the organism is, which is to say, how alive. Or perhaps it is their envy of the physicist and chemist, who have such beautiful little machines to play with — or, at least, so one may think. But it is a strange misconception, and not one shared, I suspect, by very many physicists and chemists.
One little picture can make the point. It was offered by Paul Weiss* (a cell biologist, by the way — one of the great ones of the twentieth century), and it attempts to convey something of the actual structure of molecules, as opposed to the neat stick-and-ball models every student is so familiar with. It depicts “the mapping of electron distribution in a small organic molecule”:
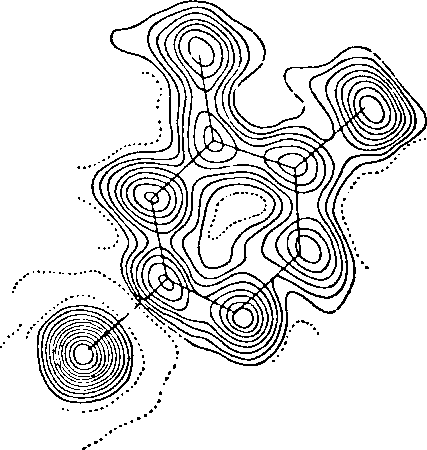
This is still a highly abstract picture, and it represents no particular things that we could readily describe as such. Perhaps the best we can do is to take the picture as schematically representing a pattern of interacting forces. We also need to imagine the “contour lines” of the drawing in fluid movement as other molecules (including water molecules) impinge upon or bind to this one. In any case, Weiss’ drawing is one every biologist would do well to meditate upon. It makes nonsense of the notion of “molecular machines”, and could have saved a lot of surprise on the part of those who are now discovering that organisms pursue their own aims in a plastic, dynamic, and well-directed manner all the way down to the molecular level.
Hilser, Vincent J. (2013). “Structured Biology: Signalling from Disordered Proteins”, Nature vol. 498 (June 20), pp. 308-10. doi:10.1038/498308a
Taylor, Susan S., Ronit Ilouz, Ping Zhang and Alexandr P. Kornev (2012). “Assembly of Allosteric Macromolecular Switches: Lessons from PKA”, Nature Reviews Molecular Cell Biology vol. 13 (Oct.), pp. 646-58. doi:10.1038/nrm3432
Weiss, Paul (1971). “One Plus One Does Not Equal Two”, in Within the Gates of Science and Beyond: Science in Its Cultural Commitments, pp. 213-61. New York: Hafner. Reprinted from The Neurosciences: A Study Program, edited by Gardner C. Quarton, Theodore Melnechuk and Francis O. Schmitt. Rockefeller University Press, 1967.
Further information: See also the June 7, 2013 post, “Signals and the whole organism”.This document: https://bwo.life/org/comm/ar/2013/are-disordered-proteins-really-disordered_5.htm
Steve Talbott :: Are “Disordered” Proteins Really Disordered?

