Biology Worthy of Life
An experiment in revivifying biology
Genes and Organisms: Improvising the Dance of Life
Stephen L. TalbottTags: chromosome/chromatin; chromosome/dynamics; DNA; epigenetics; gene/indefinability; gene regulation; genome/organization; genome/remodeling; holism; holism/contextual; plasticity; rna; transcription
Cells caught up in an intentional whole
Chromosomes without the airbrushing
And finally: The organism knows what it is doing with its DNA

You can hardly turn around today without hearing from this or that biologist or philosopher that we have gone beyond old, narrow conceptions of genes as the makers of organisms. And ours is indeed a time of great and bracing change — change, even, that portends revolution. Yet genes are still almost universally regarded among evolutionary theorists as the true bearers of destiny within the organism, which is why “genetic” remains an entrenched, if thoroughly improper, synonym for “heritable”. In other words, the gene is still honored as the one intrinsic factor truly definitive for the life of the organism. Full implications of the fact that organisms live their lives as irreducible wholes remain largely ignored and even taboo.
The taboo is not hard to understand, since we can fully acknowledge an organism’s capacity to be an organism — its integral power to act, its agency — only by abandoning the materialism and the machine models that have captivated biologists for so long. It’s a difficult step. Faced with the question where the behavioral unity of the organism resides (Who is doing the behaving?) few are prepared to acknowledge that the organism’s coherence is a coherence of intention, idea, and reason operating at an organic level.
The word “agency” may be infiltrating the vocabulary of a few philosophers and biologists, but one guesses that they can mention the necessarily implied being, or agent, only at peril of their career.
Those rare moments in life when one can, at least as an experiment, step outside the strictures of a previously unexamined taboo and let one’s calm, objective thinking roam in once-forbidden intellectual territory can certainly prove disturbing. But they can also be profoundly freeing. I hope that the following — together with the understanding of organisms and their evolution ventured in subsequent articles — will, at least for a few readers, suggest the possibility of such an experience.
Meanwhile, here I try to provide more than enough of a concrete picture of the molecular life of the organism — and particularly the use this organism makes of its genes — to justify the bold proposals that will follow.
A bodily wisdom
Perhaps you are too cold or too hot, hungry or sated, coming down with a flu or recovering from it, lifting weights or resting, thinking hard or yielding to reverie. Perhaps you have a wound that is healing, or have just now suffered a terrible psychological shock, or are concluding an intense lecture to college students. Or perhaps not much has happened at all, except that the sun has moved from the eastern to the western horizon.
Whatever your changing circumstances, the unseen physiological consequences could hardly be more dramatic. The performances of countless cells in your body are redirected and coordinated as part of a global narrative for which no localized controller exists. This redirection and coordination includes a unique choreography of gene expression in each individual cell. Hundreds or thousands of DNA sequences move (or are moved) within vast numbers of cell nuclei, and are subjected to extraordinarily nuanced, locally modulated chemical activity so as to contribute appropriately to bodily requirements that are nowhere codified — least of all in those DNA sequences.
But let’s place before our attention a more concrete picture.
In his little book, The Directiveness of Organic Activities (published in 1945),1 British biologist E. S. Russell described contemporary work on wound healing in the blood-sucking hemipteran bug, Rhodnius prolixus. Beneath the hard, outer cuticle of this insect is a single layer of epidermal cells on top of a basement membrane. If you excise a tiny sliver of these tissues, you set in motion a remarkable series of healing processes.

Rhodnius prolixus. (Thierry Heger)
To begin with, the neighboring epidermal cells become activated and migrate toward the edges of the cut, while red blood cells accumulate in the same area beneath the basement membrane. Having congregated at the site of injury, the epidermal cells then spread into the excised area. In simple cases, where the wound is small and the basement membrane intact, the wound is quickly covered by a few cells that are spread excessively thinly, with cytoplasmic bridges connecting them. As more cells follow these, they become more and more crowded until the normal density is reached, at which point the spreading ceases. After the migration, cell division continues, but mainly in the now-thinned area from which the migrating cells came. As for the cells that spread over the cut, they initially form a layer several cells thick, but the normal one-layer-thick epidermis is slowly restored through selective degeneration of the unwanted cells in the lower layers. Any overcrowding around the margins of the wound resulting from the migration of cells is similarly relieved by the degeneration of superfluous cells.
It’s good to imagine this elaborately organized, sequential activity in detail. There can be no doubt that we are seeing a norm — the organism’s own unique wholeness and integrity — being reestablished:
The end-state or terminus towards which the process moves is the restoration of the continuity of the epidermis, the replacement of cuticle and basement membrane, the re-establishment of the normal density of nuclei — a complex result, reached through appropriate activities of cells, which are here the agents concerned. These activities are of several kinds. They are behavioural — as shown in the active migration and spreading of the epidermal cells. They are physiological, as in the secretion of new cuticle. They are “morphoplastic”, as in activation and cell division; cells also degenerate where they are superfluous or unwanted.
Most interesting, however, is what happens when conditions are varied, and the same norm is restored, but by a very different route. For example, using heat, it’s possible to destroy a group of epidermal cells without injury to the overlying cuticle. In this case there is little migration toward the burn margin from surrounding areas. Rather, the existing cells at the immediate margin begin to fill in over the layer of burned cells — and they do so through multiplication within this zone of spreading rather than through migration from the periphery.
Compare this with the incision, where the injured area was filled to "overcrowding" by migration, with subsequent die-off of excess cells in the injured area. And whereas, with the incision, cell multiplication occurred in the more distant regions from which migration occurred, in the case of the burn, multiplication takes place in the injured area.
It seems that a general truth of healing processes is that they culminate, as far as possible, in the restoration of normal form and functioning. Depending on conditions, there can be a remarkable variation of means toward this end.
The point is not at all that there are no lawfully connected physical processes every step of the way, but only that the immediate causal factors are caught up in a larger pattern that governs them. No study of well-behaved local interactions shows us why those interactions should be coordinated in the plastic, goal-directed, context-sensitive manner we observe — a manner that enables them to reach the same end by different pathways, depending on the circumstances encountered.
When we look at the pattern in its own terms instead of merely adding separate physical causes together, we see a directional logic of the pattern as such, but not a necessity for any particular causal sequence.
It is, of course, a long way from the simplest possible injury of Rhodnius prolixus to a complex wound of Homo sapiens. Here is a general description of the kind of thing that goes on when you or I suffer the “assaults” of a surgeon — wounds typically of a sort that our species never before encountered during its evolutionary history.2 It comes from another British biologist, Brian Ford:
Surgery is war. It is impossible to envisage the sheer complexity of what happens within a surgical wound. It is a microscopical scene of devastation. Muscle cells have been crudely crushed, nerves ripped asunder; the scalpel blade has slashed and separated close communities of tissues, rupturing long-established networks of blood vessels. After the operation, broken and cut tissues are crushed together by the surgeon’s crude clamps. There is no circulation of blood or lymph across the suture.
Yet within seconds of the assault, the single cells are stirred into action. They use unimaginable senses to detect what has happened and start to respond. Stem cells specialize to become the spiky-looking cells of the stratum spinosum [a layer of the epidermis]; the shattered capillaries are meticulously repaired, new cells form layers of smooth muscle in the blood-vessel walls and neat endothelium; nerve fibres extend towards the site of the suture to restore the tactile senses...These phenomena require individual cells to work out what they need to do. And the ingenious restoration of the blood-vessel network reveals that there is an over-arching sense of the structure of the whole area in which this remarkable repair takes place. So too does the restoration of the skin. Cells that carry out the repair are subtly coordinated so that the skin surface, the contour of which they cannot surely detect, is restored in a form that is close to perfect.3
It is well to reflect diligently upon that phrase, “an over-arching sense of the structure of the whole area”. It is not a phrase that biologists today know what to do with. Who or what possesses this sense? And if “sense” is the wrong word, what is the right one?
Cells caught up in an intentional whole
Think concretely about that surgical wound. You’re a nearby epidermal cell, and you need to migrate. In which direction? When do you stop? And how do you reorganize all your constituent elements so as to bring yourself into movement — movement away from the place where you’ve long been content to remain settled?
Or you’re a nerve cell, and you need to participate in the extension of a nerve fiber. Again, in which direction, and by means of what sort of mobilization of all your internal processes?
Or perhaps you’re a stem cell and you need to begin a process of differentiation. But differentiation into what sort of other cell? And how do you go about a radical change in who you are? If change is going on everywhere around you, what gives anything its specific “operational advice”?
Everything needs to be accomplished in the right sequence, and in harmony with everything else going on — all this amid what looks for all the world like a chaotic disaster scene. How are we to imagine the ultimate and nearly incomprehensible coherence of the larger picture?
Now, rare is the biologist today who will hear such questions without thinking: “He is trying to suggest that there is no physical explanation adequate to these living processes. So he believes there must be some sort of vital force or miraculous guidance to make things happen”.
But this misses the mark entirely. The physical continuity of the entire scenario is unquestioned. Russell, for example, was always looking for immediate physical interactions. In Rhodnius prolixus,
observation shows that the migrating cells are specially attracted towards areas containing dead and damaged cells, and this suggests that the stimulus to activation is provided by chemical substances produced by the injured cells, and that migration towards the wound is a “chemotactic” response to these substances.
Yet Russell did not confuse this physical continuity of local interactions with what he somewhat awkwardly referred to as the “directiveness” of the larger storyline in which these interactions are caught up. It’s a confusion that biologists today almost universally consent to.
It’s not hard to observe one’s own reaction to the statement that migrating cells are activated and directed by a chemical gradient resulting from the death of nearby cells. “Oh, that explains it”. But what has happened with this “explanation”? The entire picture of cell migration — a complex and radically context-dependent mobilization of the cell that biologists have barely begun to understand — has been reduced in thought to an object here and an attractant there. It’s an almost mechanical schema — hardly problematic at all! We might as well be thinking of two rigidly interlocking gears, given that we have blocked from our minds the crucial thing: how do all the local physical interactions adaptively cohere as part of meaningful, “directive” processes, such as wound healing?
Allow me to characterize just a small part of the molecular biological reality of the organism. If a cell involved in wound healing is truly to mobilize itself, one of the many things it must do is decide how to initiate, discontinue, vary, or hold steady the expression of each one among thousands of genes. In every case there are myriad “decisions” to be made about exactly how this expression is to be managed.
The performance of a lifetime
Leaping tall edifices of thought in a single bound, we will pass over the question how cells “know” which genes need to be expressed within, say, the context of wound healing. We will also avoid asking how any given cell, a minuscule part of an overall process such as wound healing, finds its own proper place within that process. And so, assuming all the necessary contextualization to be somehow wisely taken care of, we will imagine just a single cell setting about a single task: to adjust the expression of one among its 25,000 or so genes. How might this cell proceed?
Our imaginative exercise will necessarily be more than a little artificial. That’s because we cannot help needing to think one thing at a time, whereas in the cell countless mutually entangled things are all happening at once. But we will try to make the best of it.
DNA in its larger matrix
You may recall from my earlier article, “Getting Over the Code Delusion” (Talbott 2010), that packing DNA into a typical cell nucleus is like packing about 24 miles of very thin, double-stranded string into a tennis ball, with the string cut up (in the normal human case) into 46 pieces, corresponding to our 46 chromosomes.
Box 1
A Mental Health Warning
I interrupt this narrative to bring you a public service announcement. The following descriptions contain language that ought to be disturbing, but, unfortunately, probably won’t be. I mean the routine language of molecular biology, where atomic- and molecular-level “contents” (whatever those may turn out to be in reality) are routinely described as if we were talking about so many chunks of familiar matter bumping into, or otherwise interacting with, each other. Making matters worse, the conventional language I will employ below is supplemented by sensuous, falsely airbrushed pictures, dangerous to your intellectual health.
It shouldn’t require physicists to remind us that we don’t really know what we are talking about in the domain of “particles” or particle aggregations — not, at least, if our descriptive language goes beyond the notion that some sort of play of fields, forces, and probabilities is at work. The one thing we do know is that the pictured and described stuff is not there as usually pictured and described.
The irony is that, by employing the conventional language — and I have little choice if I am to summarize the thinking of molecular biologists — I will be encouraging the very sort of causal, billiard-ball thinking that I am attempting to combat in this article.
So: I stand guilty, and you have been warned.
To locate a protein-coding gene of typical size within all that DNA is like homing in on a one-half-inch stretch within those 24 miles. Or, rather, two relevant half-inch stretches located on different pieces of string, since we typically have two copies of any given gene. Except that sometimes one copy differs from the other and one version is not supposed to be expressed, or one version needs to be expressed more than the other, or the product of one needs to be modified relative to the other. So part of the job may be to distinguish one of those half-inch stretches from the other. “Decisions” everywhere, it seems.
But no such decisions are made in a vacuum. As it happens, the chromosome does not consist of a naked DNA double helix. Our DNA, rather, is bound up with a massive, intricate, and dynamic protein-RNA-small molecule complex (called chromatin) that is as fully “informative” for the cell as the DNA sequence itself — and, you might say, much more active and directive.
Box 2
The Nucleosome
Here is one prominent feature of the larger chromosomal complex within which DNA is embedded — the nucleosome. It consists of eight core histone protein subunits enwrapped by DNA, together with a linker histone. The latter plays a role, both in influencing how the DNA is bound to the histone core, and also in managing the packing together and unpacking of neighboring nucleosomes (of which there are millions in the human genome).
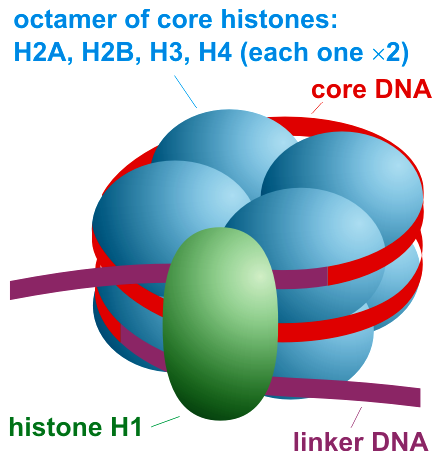
Figure 1 (credit4)
Schematically, the nucleosome is often represented as shown on the right. Eight histone proteins (blue) form the histone core particle of the nucleosome, and a linker histone (green) helps manage how the DNA is bound to the core particle. A short length of linker DNA occurs between neighboring nucleosomes.
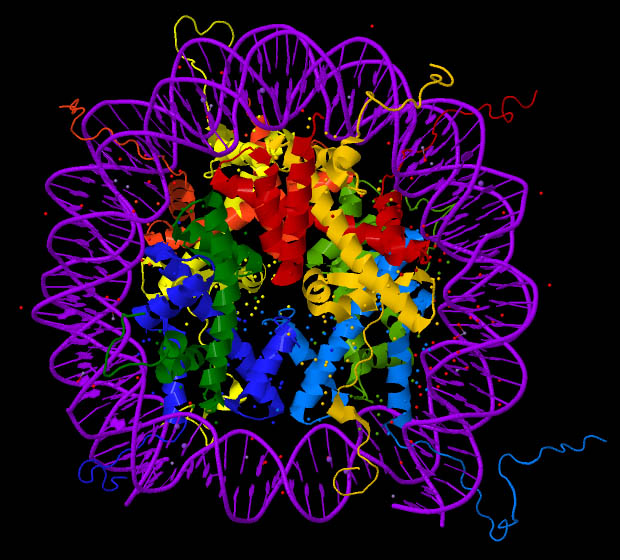
Figure 2 (credit5)
“Ribbon” images, as in Figure 2, though still highly schematic, are intended to signify some aspects of protein structure. The DNA encircling the protein histones is shown in purple. (Click on the figure in order to see an enlargement of the image.)
And yet again, though still with extreme artificiality in terms of the visual image, we have representations such as the one below, which are generated using data from sophisticated molecular imaging techniques. The red, white, and blue stick figure represents the DNA encircling (usually about one and two-thirds times) the histone core particle. Bear in mind: the “particle” shown here is really just a representation of the spatial distribution and intensity of certain forces.
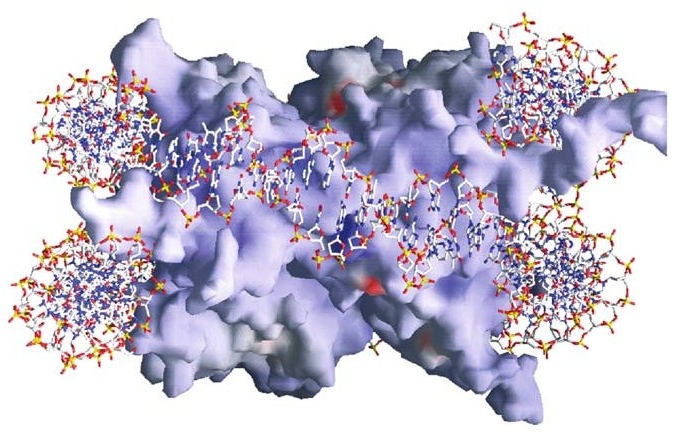
Figure 3 (credit6)
(See Box 2 for one aspect of DNA’s contextualization. We will return to the illustrations later, to indicate something of their enormous significance.)
Some of the constituents of the chromosomal “infrastructure” can bind directly to DNA and facilitate or block the transcription of this or that gene. But they can also contribute to other functions — condensing or decondensing the packing of the DNA; moving chromosomes to different regions of the cell nucleus; attaching parts of chromosomes to the nuclear envelope; interweaving and (almost miraculously, it might seem) disentangling the various chromosomes so as to form local “communities” of functionally related chromosomal loci; twisting apart the two strands of the double helix in some places and twisting them more tightly in others; altering the electrical characteristics of particular loci, and so on almost without end.
All this activity contributes to what the cell ends up producing from its DNA. For example, the loosening (untwisting) or tightening of the two strands of the double helix can make the difference between a gene’s accessibility or inaccessibility to the factors required for its transcription.
As you may surmise, then, it’s not as if any gene possessed a fixed genetic code specifying a fixed result. The result arises rather in the way a musical performance is evoked from a jazz orchestra. Like an instrument of the orchestra, a distinct locus of DNA has its own character and range of potential contributions. But there is no telling — no predicting solely from an analysis of DNA — how the locus may be employed within the improvised cellular performance.
One of many examples: the cell, by managing the shifting patterns of the chromatin infrastructure within which DNA is embedded, brings our chromosomes into movement on widely varying scales. These include large looping movements that put particular genes into connection with essential regulatory sequences and with other, related genes (that is, with other one-half inch stretches of our “24 miles of string in a tennis ball”).
Given the importance of movements such as this, how might we visualize the overall pattern and temporal narrative of gene expression? I have mentioned an orchestral performance. But perhaps we would do better to imagine an exquisitely detailed, never-ending, self-assured, yet highly improvisational dance involving billions of dancers — all coordinated with the choreography in neighboring cells and with the ongoing story of the organism as a whole. The production is a long way from that of a calculating or information-processing machine.7
Getting started
A gene is not in any case the kind of rigidly defined entity one might hope to calculate with. As a functional unit appropriate to current circumstances, it must be cobbled together by the cell according to the needs of the moment. There is no neatly predefined path to follow once the cell has located the “right” half inch or so of string, or once it has done whatever is necessary to bring that locus into proper relation with other chromosomal loci participating in the same “dance”.
One issue has to do with the fact that there are two strands in the DNA double helix and, (in a chemical sense) these complementary strands of the double helix “point” in opposite directions. In humans, protein-coding sequences can occur on both strands. Likewise, transcription (of both protein-coding and regulatory sequences) occurs on both strands, which is to say that the transcribing enzyme (RNA polymerase) can move in either direction along the double helix, depending on which strand it is straddling and what sequences exist at the current locus.
And even when the cell would proceed in one particular direction, it must “choose” the exact point in the genetic sequence at which to begin. Different starting points can yield functionally distinct results. “Many studies focusing on single genes have shown that the choice of a specific transcription start site has critical roles during development and cell differentiation, and aberrations in … transcription start site use lead to various diseases including cancer, neuropsychiatric disorders, and developmental disorders”.8
Intertwined with all the preceding issues is the task of assembling a pre-initiation complex (PIC). Figure 4 is a cartoon figure that merely names some of the PIC elements that commonly arrange themselves on DNA (shown as a black line) at any location where gene transcription is to begin. We need to imagine, at least as a beginning, that each oval is rather like the image in Figure 3, except that here each element is intimately and dynamically interacting with the patterns of force in its neighbors. (You needn’t concern yourself with abbreviations and meanings, beyond the general description I am offering now.)
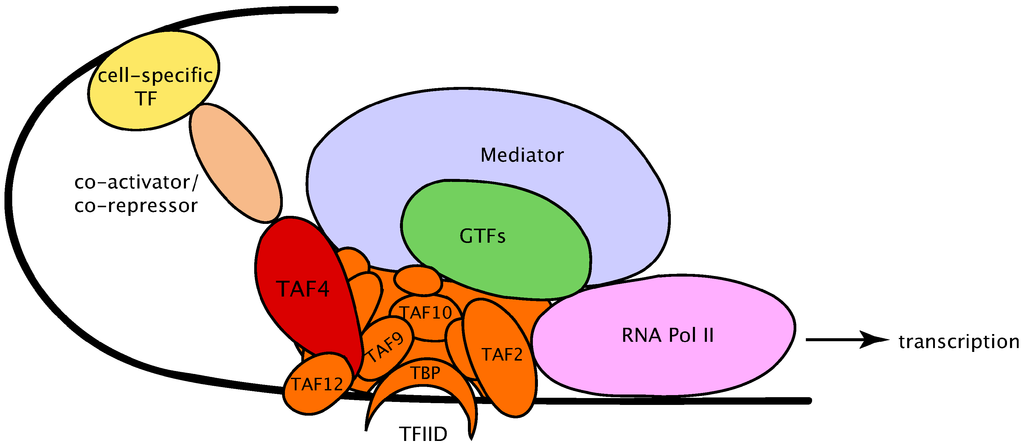
Figure 4 (credit9)
The cell’s narrative at this point could hardly be more dramatic — or more subtle. The oval named “mediator” in the drawing is a massive molecule consisting of over 30 protein subunits, arranged in various modules and interacting in numerous ways with the other PIC constituents. It varies in both structure and subunit composition, depending on context. Its effects upon gene expression are many, and still only fragmentarily grasped.
Each element of the PIC has its own story to tell. Once regarded as a mechanical, routine, and mostly unvarying assembly of “parts” whose unproblematic duty was to initiate gene transcription in a standard way, the PIC is now seen to be an infinitely varying, highly dynamic complex responding both to the immediate DNA context and to influences coming from distant reaches of the cell. Its overall “decision-making” role, which seems different from one gene to the next, is as yet hardly analyzable by researchers.
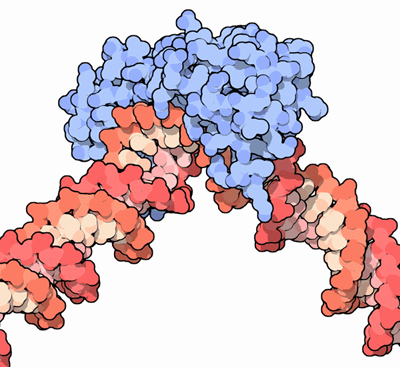
Figure 5 (credit10)
I will mention here just one other element of the pre-initiation complex. Figure 5 shows DNA in the grip of the tata-binding protein (TBP, shown in blue here, or as the crescent-moon shape at the bottom of Figure 4). The protein enters into an intricate and rather tortuous embrace of the DNA — an embrace that might remind one of the interaction between two human wrestlers.11 A severe bend of about 80 degrees is thereby applied to the double helix. This bend, which also pulls the two strands of the helix apart, is a general prerequisite for the assembly and activity of the rest of the PIC. As always, the cell is doing something sculptural, not narrowly informational in the usual sense.
Carrying on
The (protein) enzyme that transcribes DNA into RNA is RNA polymerase12. The enzyme certainly does not work alone, however, and its task is by no means cut-and-dried. To begin with, its critical interactions with various elements of the pre-initiation complex help determine whether and exactly where transcription will begin, if it is to begin at all. Then, after those “decisions” have been made, RNA polymerase moves along the double helix transcribing the sequence of genetic “letters” into the complementary sequence of an RNA.
Throughout this productive journey, which is called elongation, the RNA polymerase still keeps good and necessary company. Certain co-activators modify it during its transit of a genetic locus, and these modifications not only enable transcription elongation to begin, but also provide binding sites for yet other proteins that will cooperate throughout the transcription journey.
(This table offers some perspective on the number and variety of protein factors influencing elongation.)
I will mention here only one aspect of this cooperation of multiple factors. Transcription is an essentially rhythmical performance, with various sorts of pauses along the way. (Again, dynamic sculpture, or dance!) One pause of great significance occurs after RNA polymerase has just begun transcribing DNA but before it has fully separated from the pre-initiation complex. The factors that influence whether transcription will continue at this point — or remain paused for an extended period — play a large role in the regulation of gene expression.
But once that first pause is ended, a succession of further, generally briefer pauses continues along the way. These have to do, at least in part, with the need to disengage DNA from the frequently encountered nucleosomal core particles discussed in Box 2. The polymerase has various assistants to aid in this disengagement; they may even disassemble the core particles altogether. In the latter case the core particles will need to be reassembled behind the transcribing complex, and the nuanced meanings embodied in their composition and structure will typically have to be preserved or reestablished.
The rhythm of pauses depends, of course, on the positioning of nucleosomes along the double helix, which will vary from one gene to another and even from one time to another. All this, and not just the so-called genetic code as such, shapes the functional significance of the DNA sequence within its chromosomal context. As we will see in a moment, different proteins are produced, depending on the timing of the pauses. So it appears that the references to “choreography” and “dance” one sometimes encounters in the literature may be more than mere poetic niceties.
Shaping a significant end
Finally — and mirroring all the possibilities surrounding initiation of gene transcription — there are the issues relating to its termination. Again, they are far too many to mention here. Transcription may conclude at a more or less canonical terminus, or at an alternative terminus, or it may proceed altogether past the gene locus, even to the point of overlapping what, by usual definitions, would be regarded as a separate gene farther “downstream”. The cell has great flexibility in determining what, on any given occasion, counts as a gene, or transcriptional unit.
The last part of the transcribed gene is generally non-protein-coding, but nevertheless contains great significance. Examining this region in a single gene, a research team recently identified “at least 35 distinct regulatory elements” to which other molecules can bind.13 Further regulatory potentials arise from yet more binding sites on the customized “tail” that the cell adds to the RNA immediately upon conclusion of its transcription.
Proteins and other molecules that bind to the various regulatory elements of the non-protein-coding portion of the transcript do so in a context-sensitive manner, where cell and tissue type, phase of the cell cycle, developmental stage, location of the RNA within the cell, and environmental factors, both intra- and extra-cellular, may all play a role. These converging influences can change the stability of the RNA, change its localization within the cell, and change the efficiency of its translation into protein, among other possibilities.
But it’s not only the RNA sequence that provides opportunities for cellular management. The three-dimensional structure of the molecule offers boundless occasion for further regulation. So here, as with DNA, we find gene expression to be in part a matter of sculptural expression. And, again, it is not just a matter of static form, but of movement. “RNA dynamics play a fundamental role in many cellular functions”:
[There are] many structural maneuvers that occur over timescales ranging from picoseconds to seconds … These transitions include large-scale secondary-structural transitions at >0.1-s timescales, base pair/tertiary dynamics at microsecond-to-millisecond timescales, stacking dynamics at timescales ranging from nanoseconds to microseconds, and other ‘jittering’ motions at timescales ranging from picoseconds to nanoseconds. RNAs often harness multiple modes to achieve complex functionality.14
A drama without boundaries
Gene transcription is merely the beginning of all the activity shaping gene expression. The remaining major stages are fully as rich and multifaceted as the stages discussed above. But, with painful brevity, I will conclude this overview with just a few bare allusions. Then we will look, briefly again, at what reveals itself when we apply successively stronger magnifying glasses in order to “home in” on a single detail of the overall picture.
What is generally considered the post-transcriptional modulation of gene expression actually begins during transcription proper. A prime example has to do with what happens partly as a result of the pauses during elongation.
Cells don’t just passively accept the RNAs that emerge from the transcription process, but rather “snip and stitch” them via an elaborate procedure known as RNA splicing. It happens that the cutting out and knitting together of selected pieces typically begins before the RNA is fully transcribed, and the rhythm of pauses during elongation has an important influence upon which pieces form the mature transcript.
This splicing operation, which is applied to nearly all human RNAs, is performed by the spliceosome, consisting of a few non-protein-coding RNAs and over 300 cooperating proteins, and is hardly less exacting in its requirements than, say, brain surgery.
For the vast majority of human genes the operation can be performed in different ways, yielding distinct proteins (called isoforms) from a single RNA derived from a single DNA sequence. This is called alternative splicing, and it would be hard to find anything in human development, disease etiology, or normal functioning that is not dependent in one way or another on the effectiveness of this liberty the cell takes with its gene products.
But RNA splicing is hardly the end of it. Through RNA editing the cell can add, delete, or substitute individual “letters” of the RNA sequence.15 Or, leaving the letters in place, the cell can chemically modify them in any of over one hundred different ways.16
A whole different area of research, massive in scale, involves small, non-protein-coding RNAs, such as microRNAs (miRNAs). These target great numbers of protein-coding RNAs, leading to their degradation (which may be crucial under changing conditions) or otherwise tuning their amounts and functions.
Such degradation is an example of RNA decay in general, for which there are many different, interwoven pathways in cells. It is easy to overlook the fact that decay is fully as important — and fully as much in need of careful regulation — as the production of the RNA in the first place. During development, for example, cell differentiation would be impossible if the RNAs and proteins appropriate for an earlier form of a cell could not be gotten rid of (recycled) in favor of the RNAs serving the forthcoming, differentiated form. The same general principle holds for all changing conditions that require fresh responses from the cell.
One currently unfolding story about RNA regulation points to a complexity almost beyond all hope of grasping. Evidence now suggests that just about any RNA can help to regulate any number of other RNAs, just as it in turn is regulated by them. This is due not only to the “competition” for resources (an extremely abundant RNA might make it more difficult for less abundant ones to be translated into protein), but also to the impact of microRNAs. Many of our RNAs are densely covered with binding sequences for microRNAs, so that a typical microRNA will find about 200 different RNA species it can latch onto and degrade or otherwise affect.
This means that if a particular RNA is being highly expressed, or if it happens to be a “microRNA sponge” — that is, if it possesses multiple binding sites for a particular microRNA — it can have the effect of up-regulating other RNAs that are targets for the same microRNA. This is because it “soaks up” most of the microRNAs that might otherwise degrade those other targets. Tracking this mutual, broad-scale, and often subtle interaction where “everything seems to be affecting everything else” will presumably challenge researchers for a very long while.
Eventually, a protein-coding RNA needs to be translated into protein. This happens by means of large molecular complexes called “ribosomes”. Just as with gene transcription, there are many associated factors that must work together to bring about the initiation of translation, many that cooperate with the ribosome during translation, and yet others that play a role in modifying, localizing, or otherwise regulating the newly produced protein.
The overall picture of gene expression is one of unsurveyable complexity in the service of remarkably effective living processes. In subsequent articles I will comment on the implications of this picture for our understanding of organisms. But to continue our story here, I will now try to fashion a viewport through which we can quickly scan an entire chromosome and then “burrow down” in ever finer detail until we are confronting a minuscule part of the whole.
Chromosomes without the airbrushing
Since Francis Crick and James Watson’s elucidation of the structure of DNA in 1953, biologists have been “in denial”, according to Nature columnist, Philip Ball. “That beautiful double helix, with its genetic information written into the spiral staircase of paired nucleic-acid bases, offers such an elegant picture of the chemical principles of life and inheritance that everyone fell for it”.
The spiraling image Ball refers to has become a dominant icon of the modern era, channeling the imagination along the alluring lines of its own geometric perfection. Yet its ubiquity and influence is matched only by its falsehood. For “when we come face to face with DNA in the cell”, writes Ball, “it’s like meeting a movie star whose airbrushed publicity photos don’t look at all like the real thing. You would barely recognise Crick and Watson’s perfectly-formed molecule in the tangled, twisted and bent spaghetti that is stuffed inside the nuclei of our cells”.17
“Stuffed” probably isn’t the right word, given the functional sophistication of the ever-shifting sculptural forms whose activity in the nucleus “speaks” with such effective eloquence and on so many different levels. But let us see what we can of this “twisted and bent spaghetti”.
First look. The functioning of chromatin, the substance of chromosomes, is a matter, not of the serene, immaterial logic of an immortal code, but of a life-sustaining narrative performance. Think, for example, of the drama of mitotic cell division: chromosomes, previously replicated, become condensed some fifty-fold, are organized on the mitotic spindle through a delicate balance of tensions, and then are properly apportioned into the two daughter cells. Think, that is, what it might take to accomplish this.
But at every phase of the cell cycle the drama continues, and likewise through every stage of cell differentiation during an organism’s development. Similarly during illness, or high levels of emotional stress, or extreme exercise, or wound healing, or the passage from day to night and from night to day — in all the circumstances of life our chromosomes are gesturing their functions. That is, their movements and spatial configurations are integral to their functional roles within specific contexts. They “speak” gesturally, not merely through a passive and unvarying, computer-like configuration of hardware “logic gates”.
The performance we are looking at includes profound transformations of the double helix. For example, millions of “letters” in the DNA sequence are chemically altered, temporarily or for the longer term, by a process known as DNA methylation. By changing the local physical properties of the double helix, this modification “is observed to either inhibit or facilitate [DNA] strand separation, depending on methylation level and sequence context”.18 This has a direct effect on gene expression, since strand separation is essential for the work of the enzyme that transcribes DNA.
Moreover, DNA methylation is considered a primary instrument of cellular memory, yet even where methylation levels are more or less stably sustained, the constancy is achieved through continual exchange of methyl groups. The average half-life of a particular methyl group on any given letter is measured in hours.19 So even cellular “memory”, it would appear, is a function of dynamics rather than inert structures. In general, when it comes to chromosomes and DNA, performance, not static structure, is the fundamental reality.
The coming and going of DNA-associated proteins such as transcription factors is even more dramatic. Here the binding (to DNA or chromatin) and subsequent release can involve the cooperative or antagonistic activity of many molecules, and the temporal and spatial pattern of this activity is often what proves decisive for gene expression. Referring to “time-dependent events” as the important “‘fourth dimension’ of transcriptional regulation”, Bryan Turner, a geneticist at the University of Birmingham in the UK, notes that “studies of site-specific [transcription] factor binding in individual living cells have revealed residence times on chromatin measured in seconds”.20
All this makes for an extraordinarily complex and dynamic picture, particularly when you consider this: looking at a single gene (Gal1 in baker’s yeast) at a particular moment, researchers recently identified more than fifty proteins residing at its locus.21
In sum, there is “a surprisingly high mobility of proteins in the nucleus”.22 Oscillations, rhythms, critical rates, continual exchange of subunits — these are now known to be common features of many molecular complexes or “structures” once thought to be more or less static. The architecture of the cell nucleus sometimes appears to be compounded more of standing waves than of rigid structures. Things get done by means of a startlingly indeterminate fluidity, not the huffing and puffing of imaginary “molecular machines”.
“It was amusing”, writes cell biologist Thoru Pederson of the University of Massachusetts Medical School, “to recall the incredulity expressed by some that … chromosomes, relatively giant structures, are moving” — and doing so in the normal course of their business, even when the cell is not dividing. And yet, he continues, what else should we have expected?23
Is it such a surprise, after all, to find that the processes of life are … living processes? The idea that the essence of living things lay in a crystalline genetic code never had anything going for it apart from the mind’s preference for neat logical necessity (abstract and self-contained “explanation”) over observation and the reading of gestural narrative.
Second look. Focusing our vision more narrowly on one element of chromatin, we can home in on the millions of nucleosomes. I began my description of gene expression by mentioning the packing of chromosomes into the cell nucleus, and (in Box 2) said a little about the role of the nucleosome in this packing and in gene regulation generally. The nucleosomal core particle we should now note, “is much more flexible than the crystal structure [which is the basis for images like Figures 2 and 3] might lead us to believe”, and our current understanding of it “does not lend itself to simplifying generalisations”.24
In particular, nucleosomes are partially or wholly disassembled during gene transcription, and then reassembled after the transcribing enzyme (RNA polymerase) has passed by. However, the rate of this assembly and disassembly differs with different genes, and has been proposed as one of the factors regulating gene expression.
Beyond that, individual histones of the nucleosome (there are eight in a fully constituted core particle) can come and go at an almost alarming rate — with an average exchange rate of just a few minutes for many nucleosomes, especially those associated with active genes. And the histones exchanged in this way can be different histones — histone variants — with each variant exerting its own sort of influence on gene expression and chromatin dynamics.
Like DNA methylation, histone variants are associated with the formation of cellular memory. (The members of a cell lineage, beginning with a stem cell, must proceed through a series of cell divisions on the way, for example, to a pancreatic islet cell destination. Each cell along the path needs to “remember” the trajectory along which it has been traveling — a trajectory leading toward islet cells rather than, say, muscle cells.) The puzzling discordance between the instability of histone variants, on the one hand, and their role in memory formation, on the other, is, according to University of Georgia molecular biologist Richard Meagher, “extreme”.25
At least it is if we imagine cellular memory to be like a text safely stowed away somewhere. But the organism seems to have a much more complex and dynamic notion of memory. It is well to keep in mind that, whatever we may mean by a cell’s “remembering” where it is coming from, it must also be continually departing from the ways of its ancestors in order to reach where it is getting to. The story is through and through one of activity and transformation. The character of that activity within its larger active context, and not any fixed structure, may be the more essential bearer of identity and meaning.
One of the many aspects of the relation between the nucleosomal core particle and the encircling DNA has to do with the electrical attraction or repulsion between each local region of the core particle and the associated DNA. Also significant is which part of the DNA double helix at any point faces outward from the histones and which part faces inward. These factors directly affect the accessibility of any given DNA sequence to transcription factors and other regulatory molecules.
The nucleosome, we can fairly say, is a ceaselessly transforming matrix and organizational hub whose structure and pattern of activity is never exactly duplicated anywhere in the genome, nor ever sustained unchanged from one minute to the next. It is where the infinitely ramified interface between the larger cell and its DNA comes to its most focal expression. And that expression turns out to be livingly nuanced activity, dynamic beyond what anyone imagined.
Third look. We will burrow still further down into chromatin. I present below a modestly enlarged view of Figure 2, showing a ribbon diagram of a nucleosome — a histone core particle with the DNA double helix (in purple) wrapped around it a couple of times. You will note a number of squiggly “pig’s tails” extending outward from the core histones. These are the thin, flexible, and mobile histone tails. There are hundreds of distinct chemical modifications of these tails (referred to as post-translational modifications, or PTMs), and the countless resulting patterns within any given nucleosome or group of nucleosomes are intimately bound up with the expression of genes. In fact, there is virtually nothing relating to gene regulation, DNA replication, chromatin structure and dynamics, or the overall functional organization of the nucleus that is not correlated in one way or another with patterns of histone tail modifications.

Figure 4. An enlarged view of Figure 2, showing a ribbon drawing of a nucleosome.
Learning about these tails, we are — albeit across a considerable conceptual divide — easily reminded of both the “sensory” functions of insect antennae and the “motor” functions of limbs. On one hand, the tails are receivers of signals coming from all quarters and provide a context where the integrated significance of the signals can be “read off” (to use the standard phrase) by the innumerable gene-regulatory proteins that are sensitive to them. In this way, histone tail modifications, or combinations of them, “recruit” (again the standard phrase) various proteins that either restructure chromatin in one way or another, or more directly regulate the expression of particular genes.
There is in fact a galaxy of proteins that interact with single modifications, or with groups of them, or with the asymmetrically modified tails of a histone pair, or with a histone modification in proximity to a site of DNA methylation. Every such protein acts out of its own world of biochemical genesis, modification, and protein regulation, and together these proteins tell a great part of the story of gene regulation. Unfortunately, I can say almost nothing about them here.
Apart from their “sensory” function, the mobile tails can insinuate themselves into one of the grooves of the double helix, thereby loosening the DNA from the nucleosomal core particle (and making it more available for transcription), or else binding it more tightly. In both cases, one way this is accomplished is by altering the electrical balance between histone and DNA.
But a tail can also interact with the globular core of its own or a different histone, and can even find an affinity with an altogether different nucleosome, thereby having a potentially huge effect on the packing of the local chromatin. This, again, can make genes either more or less accessible for transcription and various forms of regulation.
Perhaps you can now see why the members of one research team, writing about histone tail modifications, find themselves reflecting upon
the incredibly intricate nature of the chromatin landscape and resultant interactions. The biological consequences of [interactions between histone tail modifications and regulatory proteins] are highly context dependent, relying on the combinatorial readout of the spatially and temporally fluctuating local [chromatin] environment and leading to a highly fine-tuned [regulation] of particular genomic sites.26
Fourth look.
Magnifying our view one last time, we will home in a single histone tail modification. I choose the one called ubiquitination simply because its gene regulatory roles do not seem quite as extensive as those performed by some of the other tail modifications — or, at least, its roles have not been as extensively documented. This makes their description here a little more manageable.
Monoubiquitination is the “attachment” (a poor word) of a single ubiquitin chemical group to a lysine amino acid of a protein. In the case of the histone tails, this can be done at more than one lysine, but we will look only at the monoubiquitination of lysine 120 on the tail of the histone known as H2B, all of which can be referred to as H2BK120ub1 (abbreviated here as H2Bub1).
So what is the significance of this modification at a single histone tail location? Here’s one summary:
H2Bub1 takes part in almost every molecular process associated with chromatin biology. H2Bub1 has been shown to regulate transcription initiation and elongation, DNA damage response and repair, DNA replication, nucleosome positioning, RNA processing and export, chromatin segregation and maintenance of chromatin boundaries. Given the large number of molecular processes regulated by H2Bub1, it is not surprising that H2Bub1 plays a vital role in some of the most fundamental biological processes that occur within multicellular organisms. [Loss of an enzyme responsible for ubiquitination] results in very early embryonic lethality. Furthermore, aberrant H2Bub1 levels can affect cell cycle progression, apoptosis [“programmed cell death”], stem cell differentiation, development, viral infection outcome and tumorigenesis.27
Of course, H2Bub1 does nothing “in general”; specific results are always context-dependent. For example, blocking this modification in a particular human cell line was found to upregulate some genes, downregulate others, and leave a great many unchanged. Under some circumstances, H2Bub1 is particularly needed for the transcription of relatively long genes. And the modification also plays an important role in histone “crosstalk”, helping to regulate other crucial modifications within the same or on different histones.
A search for “effector” molecules that, singly or cooperatively, associate and interact with (“read”) the H2Bub1 modification led to the identification of more than ninety proteins, many with known functions in gene regulation consistent with those of H2Bub1. This points us to what could be our “fifth look”, analyzing one or more of those proteins, the modifications they undergo, and the larger regulatory world in which they are caught up. But there would be no end of this, whereas there most definitely is an end to readers’ patience.
It remains to mention only that, with ubiquitination, as with so many other molecular biological investigations, researchers are vexed by “a need to establish causality more unequivocally”28 — a need that never seems to be fully satisfied as understanding grows. The search for unambiguous causes is a fruitless one.29 The kind of causes being looked for don’t exist in organisms.
And finally: The organism knows what it is doing with its DNA
A decisive problem for the classical view of DNA is that “as cells differentiate and respond to stimuli in the human body, over one million different proteins are likely to be produced from less than 25,000 genes”.30 Functionally, in other words, you might say that we have over a million genes. But here the word “gene” cannot refer to a defined sequence of genetic “letters”. It must refer, in the first instance, to certain characteristic, context-dependent activities of cell and organism — activities in which DNA figures along with innumerable other players.
Box 3
The Genome in Dynamic Nuclear Space
A few comments from the literature:
![]() “The dynamic spatial organization of the
nucleus
both reflects and shapes
genome
function … We now have a picture of a
genome
that is ’structured’, not in a rigid three-dimensional network, but in a
dynamic organization [that] clearly changes during normal
development
and differentiation”.31
“The dynamic spatial organization of the
nucleus
both reflects and shapes
genome
function … We now have a picture of a
genome
that is ’structured’, not in a rigid three-dimensional network, but in a
dynamic organization [that] clearly changes during normal
development
and differentiation”.31
![]() Researches have revealed “the astounding degree to which our
genome
… appears to be dynamically utilized for the purposes of
gene
regulation”.32 Of course, the question most
immediately implied doesn’t get asked: utilized by whom, or by what?
Researches have revealed “the astounding degree to which our
genome
… appears to be dynamically utilized for the purposes of
gene
regulation”.32 Of course, the question most
immediately implied doesn’t get asked: utilized by whom, or by what?
![]() Although the researchers’ first impulse was to find in
chromatin
modifications (such as
histone tail modifications)
another “simple code”, it eventually became evident, according to
geneticist Shelley Berger of Philadelphia’s Wistar Institute, that “a more
likely model is of a sophisticated, nuanced
chromatin
‘language’ in which different combinations of basic building blocks yield
dynamic functional outcomes”.33
Although the researchers’ first impulse was to find in
chromatin
modifications (such as
histone tail modifications)
another “simple code”, it eventually became evident, according to
geneticist Shelley Berger of Philadelphia’s Wistar Institute, that “a more
likely model is of a sophisticated, nuanced
chromatin
‘language’ in which different combinations of basic building blocks yield
dynamic functional outcomes”.33
“What was previously known as junk DNA in fact appears a regulatory jungle. In order to understand the laws of the jungle, linear information must now be converted into spatial relationships”.34
![]() Indeed, the almost exclamatory recognition that
“Genomes
are incredibly dynamic”35 in both space and time has become
commonplace today, even if it still seems to surprise many. But the
appropriate questions have scarcely been addressed as yet. No one would
argue that
DNA
itself is “incredibly dynamic”, for it is just about the most inert
substance in the cell, at times approaching an almost crystalline state.
It is the cell as a whole that brings our
DNA
and
chromosomes
into the movement and directed activity through which they are made to
serve the needs of digestion, muscular exertion, sensory perception, and
all our other biological functions.
Indeed, the almost exclamatory recognition that
“Genomes
are incredibly dynamic”35 in both space and time has become
commonplace today, even if it still seems to surprise many. But the
appropriate questions have scarcely been addressed as yet. No one would
argue that
DNA
itself is “incredibly dynamic”, for it is just about the most inert
substance in the cell, at times approaching an almost crystalline state.
It is the cell as a whole that brings our
DNA
and
chromosomes
into the movement and directed activity through which they are made to
serve the needs of digestion, muscular exertion, sensory perception, and
all our other biological functions.
![]() “The
sequence
of our
genes
are [sic] like the keys on the piano; it is the context that makes the
music”.36
Except that the raw
sequence
does not even contain all the keys; let’s say: just the white keys. The
flats and sharps, without which the music would lose its savor, are
provided by
DNA methylation,
RNA editing,
and more.
“The
sequence
of our
genes
are [sic] like the keys on the piano; it is the context that makes the
music”.36
Except that the raw
sequence
does not even contain all the keys; let’s say: just the white keys. The
flats and sharps, without which the music would lose its savor, are
provided by
DNA methylation,
RNA editing,
and more.
That fairly well sums up all the foregoing. But I need, finally, to say something, not about how the organism manages the contextualization and expression of its DNA, so crucial for genetic function, but rather about its ability to alter (“mutate”) the genomic sequence itself. For it is clearly capable of inserting new sequences in DNA, deleting old ones, moving them from here to there, exchanging them between chromosomes, and so on.
Even when an unwelcome external agent such as radiation or a toxin damages DNA, our cells may not suffer the damage passively They often undertake their own reworking of the affected loci — a reworking that may extend to a thousand “letters” and more.
Further, every cell frequently initiates single- and double-strand breaks, then stitching things back together in one way or another — a frequent enough requirement, if only to facilitate the organization, disentanglement, and proper physical characteristics (such as the degree of double-stranded “twist”) of those “24 miles of string divided into 46 pieces inside a tennis ball”. And in countless immune cells, portions of the genome are intentionally “scrambled” into distinct patterns, enabling the cells to produce millions of different antibodies.
Sometimes individual genes or sections of a chromosome are duplicated in certain cells. But genome remodeling goes beyond this. Megakaryocytes (cells involved in platelet production in bone marrow) have up to 128 copies of the entire genome; hepatocytes (liver cells constituting some 3/4 of the liver’s mass) typically have 4 to 8 copies; trophoblast giant cells in the embryonic outer layer may have up to 1000 copies; and cardiomyocytes (heart muscle cells) usually have 4 copies of the genome. In some cell types such as skeletal muscles, there are many separate nuclei in a single, very large cell, with each nucleus possessing a full complement of DNA.
The still-routine statement (I have sometimes acquiesced in it myself) that “all the cells in our body have the same DNA” has been found to fall further and further from the truth, even when one neglects (as one should not) the chemical and physical DNA transformations of the sort I have discussed in earlier sections of this article. According to a recent paper, “perhaps the quantity of nuclear DNA content in human cells is best viewed as a distribution of values” rather than as a single value. New analyses are suggesting that “systematic variation in nuclear DNA content is a more ubiquitous phenomenon in human cells than was previously appreciated”.37
MIT science historian, Evelyn Fox Keller, has nicely summarized aspects of mutation research:
The regulation of mutability has become one of the hottest topics in molecular biology … But with every step toward elucidation, the picture is rendered ever more complex by the increasing wealth of detail. Scores of proteins have already been implicated in the cell’s response mechanisms, and reports of new players appear with every passing month …
Stability and mutability are proving to be flip sides of one another in the specific mechanisms by which they are controlled. Both are at the mercy of enzymatic processes, and apparently equally so. Moreover, not only are the mechanisms controlling stability and mutability held in a delicate balance, but that very balance is under cellular regulation, and it shifts in response to the particular environment in which the cell finds itself.38
Let me then state the overall lesson clearly: the organism knows what it is doing with its DNA, as with all its molecular activities. No one remodels complex, dynamic structures on the fly, employing them in meaningful and ever-changing performances, without having gained mastery over the materials he is working with.39
Yet this living, active, and governing wisdom that we confront face to face in every organism seems to threaten a kind of theoretical paralysis in biologists, who have therefore long since learned to ignore it as they pass by, whistling innocently. In subsequent articles we will try to face up to the character of the organism, and its evolution.
Notes
1. Russell 1945, pp. 12-15.
2. Actually, most any wound an animal receives involves a precise location, a precise sort of damage, and a precise distribution of damage that can hardly have been duplicated even once during evolutionary history.
4. Image by Darekk2 (Own work)
6. Image from Luger 2006.
7. This is a bit redundant, since an information-processing machine just is a kind of calculating machine.
9. Image from Roy and Singer 2015.
10. Image courtesy of David S. Goodsell and RCSB Protein Data Bank.
11. The Wikipedia article, “Tata-binding protein”, offers a succinct description of part of this interaction: “When TBP binds to a [special sequence] within the DNA, it distorts the DNA by inserting amino acid side-chains between base pairs, partially unwinding the helix, and doubly kinking it. The distortion is accomplished through a great amount of surface contact between the protein and DNA. TBP binds with the negatively charged phosphates in the DNA backbone through positively charged lysine and arginine amino acid residues. The sharp bend in the DNA is produced through projection of four bulky phenylalanine residues into the minor groove. As the DNA bends, its contact with TBP increases, thus enhancing the DNA-protein interaction”.
12. There are actually three RNA polymerase enzymes in humans: RNA polymerase I, II, and III. I will be speaking of RNA polymerase II, which transcribes the great majority of our genes. Also, “RNA” in the following descriptions will refer to messenger RNA (mRNA), which can be translated into protein, or else to RNA in general. References to specific non-protein-coding RNAs such as microRNAs (miRNAs) will be flagged as such.
13. Kristjánsdóttir, Fogarty and Grimson 2015.
14. Mustoe, Brooks and Al-Hashimi 2014.
15. Just about any functional significance of an RNA — from what protein it produces, to its stability and and cellular localization, to the various roles of its three-dimensional structure — can be affected by this editing. One kind of editing (known as A-to-I editing) “is extremely abundant in primates: over a hundred million editing sites exist in their genomes” (Levanon and Eisenberg 2014). However, biologists have only begun to explore the functional significance of most of this editing, and there remains among the majority of researchers today a tendency to dismiss as “random noise” whatever their current methods and concepts cannot helpfully illuminate.
16. Regarding one of these modifications, known as mRNA adenosine methylation (m6A), Timothy Nilsen, a molecular biologist at Case Western Reserve University in Cleveland, writes:
A series of papers have appeared in rapid succession, together providing a wealth of unequivocal evidence for m6A function. But these findings still have not led to a coherent picture of the number and variety of functions of the m6A modification. (Nilsen 2014).
18. Severin, Zou, Gaub and Schulten 2011.
21. Byrum 2013.
22. Cardoso, Schneider, Martin and Leonhardt 2012.
26. Musselman, Lalonde, Côte and Kutaleladze 2012.
27. Fuchs and Oren 2014. I draw on these authors for the information in this section.
30. de Almeida and Carmo-Fonseca 2012.
32. Joanna Wysocka in Dekker, Wysocka, Mattaj et al. 2013.
34. Splinter and de Laat 2011. The phrase “What was previously known as junk DNA”, used by Splinter and de Laat, will rouse the indignation of a certain group of evolutionary biologists who, provoked by widespread dismissals of the notion of junk DNA, have gone on the warpath. I hope to write about this controversy before long.
37. Gillooly, Hein and Damiani 2015.
38. Keller 2000, pp. 34-5. The picture Keller paints has only become more vivid during the fifteen years since publication of her book. For example, in sequencing thirty-six single neurons from the human prefrontal cortex, researchers recently found more than a thousand (and “as many as 1458 to 1580”) single-nucleotide somatic mutations per cell, “the majority of which were unique to that cell”:
Although most mutations were intergenic, some occurred in coding regions … Remarkably, a quick calculation shows that it’s likely that every coding nucleotide of every gene is mutated somewhere in the brain … [An analysis] revealed a nested pattern across the brain, exactly as would be expected of mutations that accumulate during embryonic development. Even within a few millimeters of prefrontal cortex, multiple lineages were intermingled and were also found in heart, lung, and other peripheral organs, showing that lineages must have become physically mixed in the early embryo. Indeed, any given neuron in the prefrontal cortex was more closely related [genetically] to a cardiomyocyte than to 75% of its neighboring neurons. (Linnarsson 2015; Lodato, Woodworth, Lee et al. 2015)
As an aside: this sort of work reveals, according to the author, that “the earliest cells [of the embryo] contribute to all tissues, but in highly skewed proportions”. It’s a nice picture of the integral unity of the organism. Of course, mutations are still treated almost solely as “accidents” and as the results of “failed repair”. This, however, is slowly changing. I have little doubt that it will eventually have radically changed.
Also, it’s worth bearing in mind that one thousand single-nucleotide mutations in a cell represents roughly one mutation per every three million nucleotide bases, or “letters”, of the haploid human genome. However, the researchers did not report on the lengthier mutations, such as copy-number variations and retrotransposon insertions, already known to occur in the brain.
39. University of Chicago microbiologist and geneticist, James Shapiro, has “written the book” on what he calls natural genetic engineering. See especially Shapiro 2014. Earlier treatments include Shapiro 2010 and Shapiro 2009. See also his immensely valuable book, Evolution: A View from the 21st Century (2011) and an associated website: https://shapiro.bsd.uchicago.edu.
Sources:
Ball, Philip (2008). “Pulling Our Strings”, Chemistry World (May). Available at https://www.chemistryworld.com/section/feature/pulling-our-strings/3004744.article
Berger, Shelley L. (2007). “The Complex Language of Chromatin Regulation during Transcription”, Nature vol. 447 (May 24), pp. 407-12. doi:10.1038/nature05915
Bissell, Mina (2011). “Why Don’t We Get More Cancer? A Proposed Role of the Microenvironment in Restraining Cancer Progression”, Nature Medicine vol. 17, no. 3 (March), pp. 320-9. doi:10.1038/nm.2328
Byrum, Stephanie D., Sean D. Taverna and Alan J. Tackett (2013). “Purification of a Specific Native Genomic Locus for Proteomic Analysis”, Nucleic Acids Research, vol. 41, no. 20 (e195). doi:10.1093/nar/gkt822
Cardoso, M. Cristina, Katrin Schneider, Robert M. Martin and Heinrich Leonhardt (2012). “Structure, Function and Dynamics of Nuclear Subcompartments”, Current Opinion in Cell Biology vol. 24, pp. 79-85. doi:10.1016/j.ceb.2011.12.009
Chalker, Douglas L. and Meng-Chao Yao (2011). “DNA Elimination in Ciliates: Transposon Domestication and Genome Surveillance”, Annual Review of Genetics vol. 45, pp. 227-46. doi:10.1146/annurev-genet-110410-132432
de Almeida, Sérgio F. and Maria Carmo-Fonseca (2012). “Design Principles of Interconnections Between Chromatin and pre-mRNA Splicing”, Trends in Biochemical Sciences vol. 37, no 6 (June), pp. 248–53. doi:10.1016/j.tibs.2012.02.002
Dekker, Job, Joanna Wysocka, Iain Mattaj et al. (2013). “Nuclear Biology: What’s Been Most Surprising?”, Cell vol. 152 (March 14), pp. 1207-8. doi:10.1016/j.cell.2013.02.041
Ford, Brian J. (2009). “On Intelligence in Cells: The Case for Whole Cell Biology”, Interdisciplinary Science Reviews vol. 34, no. 4 (Dec.), pp. 350-65. doi:10.1179/030801809X12529269201282
Fraser, Peter and Wendy Bickmore (2007). “Nuclear Organization of the Genome and the Potential for Gene Regulation”, Nature vol. 447 (May 24), pp. 413-7. doi:10.1038/nature05916
Fuchs, Gilad and Moshe Oren (2014). “Writing and Reading H2B Monoubiquitylation”, Biochimica et Biophysica Acta vol. 1839, pp. 694–701. doi:10.1016/j.bbagrm.2014.01.002.
Gillooly, James F., Andrew Hein and Rachel Damiani (2015). “Nuclear DNA Content Varies with Cell Size across Human Cell Types”, Cold Spring Harbor Perspectives in Biology 2015;7:a019091 (July). doi:10.1101/cshperspect.a019091
Keller, Evelyn Fox (2000). The Century of the Gene. Cambridge MA: Harvard University Press.
Klerk, Eleonora de and Peter A. C. ’t Hoen (2015). “Alternative mRNA Transcription, Processing, and Translation: Insights from RNA Sequencing”, Trends in Genetics vol. 31, no. 3 (Mar.), pp. 128-39. doi:10.1016/j.tig.2015.01.001
Kristjánsdóttir, Katla, Elizabeth A. Fogarty and Andrew Grimson (2015). “Systematic Analysis of the Hmga2 3’ UTR Identifies Many Independent Regulatory Sequences and a Novel Interaction Between Distal Sites”, RNA vol. 21, no. 7 (July), pp. 1346-60. https://www.rnajournal.org/cgi/doi/10.1261/rna.051177.115
Levanon, Erez Y. and Eli Eisenberg (2014). “Does RNA Editing Compensate for Alu Invasion of the Primate Genome?”, Bioessays vol. 37, pp. 175–81. doi:10.1002/bies.201400163
Linnarsson, Sten (2015). “A Tree of the Human Brain”, Science vol. 350, no. 6256 (Oct. 2), p. 37. doi:10.1126/science.aad2792
Lodato, Michael A., Mollie B. Woodworth, Semin Lee et al. (2015). “Somatic Mutation in Single Human Neurons Tracks Developmental and Transcriptional History”, Science vol. 350, no. 6256 (Oct. 2), pp. 94-8. doi:10.1126/science.aab1785
Luger, Karolin (2006). "Dynamic Nucleosomes", Chromosome Research vol. 14, pp. 5-16. doi:10.1007/s10577-005-1026-1
Meagher, Richard B. (2014). “‘Memory and Molecular Turnover,’ 30 Years after Inception”, Epigenetics and Chromatin vol. 7, no. 37. doi:10.1186/1756-8935-7-37
Musselman, Catherine A., Marie-Eve Lalonde, Jacques Côte and Tatiana G. Kutaleladze (2012). “Perceiving the Epigenetic Landscape Through Histone Readers”, Nature Structural and Molecular Biology vol. 19, no. 12 (Dec.), pp. 1218-27. doi:10.1038/nsmb.2436
Mustoe, Anthony M., Charles L. Brooks and Hashim M. Al-Hashimi (2014). “Hierarchy of RNA Functional Dynamics”, Annual Review of Biochemistry vol. 83, pp. 441–66. doi:10.1146/annurev-biochem-060713-035524
Nilsen, Timothy W. (2014). “Internal mRNA Methylation Finally Finds Functions”, Science vol. 343 (Mar. 14), pp. 1207-8. doi:10.1126/science.1249340
Pederson, Thoru (2011). “The Nucleus Introduced”, Cold Spring Harbor Perspectives in Biology 2011;3:a000521. doi:10.1101/cshperspect.a000521
Roy, Ananda L. and Dinah S. Singer (2015). “Core Promoters in Transcription: Old Problem, New Insights”, Trends in Biochemical Sciences vol. 40, no. 3 (Mar.), pp. 165-71. doi:10.1016/j.tibs.2015.01.007
Russell, E. S. (1945). The Directiveness of Organic Activities. Cambridge UK: Cambridge University Press.
Severin, Philip M. D., Xueqing Zou, Hermann E. Gaub and Klaus Schulten (2011). “Cytosine Methylation Alters DNA Mechanical Properties”, Nucleic Acids Research vol. 39, no. 20, pp. 8740-51. doi:10.1093/nar/gkr578
Shapiro, James A. (2009). “Revisiting the Central Dogma in the 21st Century”, Annals of the New York Academy of Sciences vol. 1178, pp. 6-28. doi:10.1111/j.1749-6632.2009.04990.x
Shapiro, James A. (2010). “Mobile DNA and Evolution in the 21st Century”, Mobile DNA vol. 1, no. 4. doi:10.1186/1759-8753-1-4
Shapiro, James A. (2011). Evolution: A View from the 21st Century. Upper Saddle River NJ: FT Press Science. There is a website associated with the book, containing additional materials: https://shapiro.bsd.uchicago.edu/evolution21.shtml
Shapiro, James A. (2014). “Physiology of the Read-Write Genome”, Journal of Physiology vol. 592, no. 11 (June 1), pp. 2319-41. doi:10.1113/jphysiol.2014.271130
Splinter, Erik and Wouter de Laat (2011). “The Complex Transcription Regulatory Landscape of Our Genome: Control in Three Dimensions”, EMBO Journal vol. 30, no. 21, pp. 4345-55. doi:10.1038/emboj.2011.344
Talbott, Stephen L. (2010). “Getting Over the Code Delusion”, The New Atlantis no. 28 (summer), pp. 3-27. Original version published in NetFuture #179 (Feb. 18, 2010). Available at http://BiologyWorthyofLife.org/mqual/genome_4.htm.
Talbott, Stephen L. (2011). “From Physical Causes to Organisms of Meaning”, The New Atlantis no. 30 (winter), pp. 24-49. Original version published as “What Do Organisms Mean?” in NetFuture #182 (Feb. 22). Available at http://BiologyWorthyofLife.org/mqual/genome_6.htm
Turner, Bryan M. (2014). “Nucleosome Signalling: An Evolving Concept”, Biochimica et Biophysica Acta vol. 1839, pp. 623-6. doi:10.1016/j.bbagrm.2014.01.001
This document: https://bwo.life/org/comm/ar/2015/genes_29.htm
Steve Talbott :: Genes and Organisms: Improvising the Dance of Life

