Biology Worthy of Life
An experiment in revivifying biology
How to Unthink Epigenetics
Stephen L. TalbottTags: chromosome/chromatin; DNA/noncoding; epigenetics; explanation/and causation; gene/genocentrism; gene regulation; holism/contextual; inwardness
I suppose it’s finally become official: epigenetics is for real. Its certificate of reality has been issued by the Roadmap Epigenomics Consortium, which last month went public with a trove of reports in Nature and some of its sister publications. A news item on the Nature website summarized the import of the papers this way:1
Almost every cell in the human body has the same DNA sequence. So why is a heart cell different from a brain cell? Cells use their DNA code in different ways, depending on their jobs — just as the orchestra [in an accompanying video presentation] can perform one piece of music in many different ways. The combination of changes in gene expression in a cell is called its epigenome.
The symphony orchestra metaphor has some promise. But, as we will see, it’s use in the Nature article is not particularly apt, if only because the “one piece of music” is not the supposed DNA code, but the organism as a whole.
But you say you’ve never quite figured out what epigenetics is? Well, you’re in excellent company. Many molecular biologists seem a little confused about it, too. But the confusion is also an opportunity, since clarifying “epigenetics” demands that we undo certain damaging intellectual habits within genetics and biology generally. This article is intended as a modest contribution toward that undoing. For some relevant background on genetics and epigenetics, you might want to look at my earlier essay, “Getting Over the Code Delusion”.
Etymologically, “epigenetics” suggests something like “on top of genetics” or “added to genetics”.2 Conventionally, the word commonly refers today to “heritable changes in gene function that are not due to changes in DNA sequence” — where a “DNA sequence” is a succession of nucleotide bases constituting the “letters” of the so-called genetic code, and “heritable” applies not only to what can pass from parent organisms to their progeny, but also what can pass from any given cell to its daughter cells.
Realistically, biologists are being driven inexorably toward a definition that looks something like this: “epigenetics deals with all the ways DNA is caught up in the activity of its larger context and brought into service of the whole”. On one hand, this definition reminds us that what is “on top of” DNA is nothing less than the whole organism functioning as a whole. On the other hand, a term that threatens to encompass just about everything begins to lose its value as a special term.3
This might usefully suggest that we retire the word epigenetics as unfortunately genocentric and just get on with describing how organisms carry out their organically integrated lives — express their own character — through all their activities and at every level of observation. But such a reckoning with the lessons emerging from today’s laboratories is problematic for those many biologists who have invested their careers in the notion that DNA “expresses” the organism rather than the other way around.
So. Do you feel you’ve got a grip on “epigenetics” now? I didn’t think so. A few more words, maybe.
DNA has long been viewed as the carrier of instructions for building an organism. But no one has ever pointed to a single instruction in DNA or anywhere else in the cell. The only things to make this metaphor seem even remotely appropriate have been the biologist’s preconceptions, shaping a fierce resolve to view the organism in machine-like or computer-like terms, come what may.4 Under the influence of that resolve, DNA, as the instructional program governing the machine-organism, became a kind of First Cause and Unmoved Mover, accounting for everything essential about the organism while remaining stably itself.
Its relative (if partly imaginary) stability made DNA wonderfully trackable by the methods of molecular biology, and stable things that can be tracked experimentally were prerequisites for the biologist’s compulsive search for definitive “mechanisms”, or causes. DNA, imagined as a fixed, almost crystalline substance, became the prime cause of the living organism simply because everything else was too alive to tie down. Only the base sequence of DNA — at least when abstracted from its dynamic chemical matrix, its living chromosomal context — remained unchanged long enough for the researcher to correlate some of its features with “causal effects”.
An insistence upon fixed causes and an abhorrence of the unassayable qualities of life — these are our best guides to the conventional meanings of epigenetic. The word expresses a certain slavish deference toward the dominant gene. This was already true in 1942, when the evolutionary geneticist and embryologist, C. H. Waddington, first defined epigenetic so as to embrace the “causal mechanisms” by which “genes of the genotype bring about phenotypic [observable] effects5”.
This deference remains undiminished in the current definition referring to heritable changes in gene function that are not due to changes in DNA sequence. Genes remain at the center; the importance of other elements is measured only by their consequences for genes, whose stably heritable functioning is what really matters. Those other elements are, as the word itself reminds us, merely epigenetic — “add-ons”, or secondary modifiers, of the essential DNA.6
The standard argument among those who have had a hard time abiding the growing research emphasis on epigenetics has been precisely that the non-genetic features under study lacked DNA’s permanence and reputed causal efficacy. The truth that the living character of an organism is most fundamentally expressed in a consistent, active responsiveness — in a coherent, stable, and qualitative movement of life — was not considered. Who, after all, wants to track expressive gestures when there are convenient things we can label as “causes”?
And so those features of the cell taken to be epigenetic came to be viewed primarily as things — as mere static “marks” associated with DNA, rather than what they really are: fluent patterns of chemical transformation of DNA and of its mobile, shape-shifting matrix. The marks came to be viewed, that is, much like the punctuation or diacritical marks associated with a text. The nice thing about this was that it allowed one to keep thinking about DNA as the First Cause of the organism. The unchanging text, after all, is the main thing; its markup can easily be thought secondary and inessential. And, indeed, a common notion — no longer defensible today — was that somehow the DNA text must one-sidedly determine the marks, as opposed to the marks playing a role in establishing the meaning of DNA.7
The two primary types of mark are those referred to as DNA methylation (involving the transformation of genomic “letters” — millions of them in the case of the human genome — through the incorporation of methyl groups) and histone modifications (through which DNA-enwrapped proteins are altered in a huge variety of ways by a number of different chemical groups). In both cases the marking patterns correlate with patterns of gene expression. The dynamics of marking, along with many other epigenetic factors, signify the unique way this or that sort of cell is using its DNA. But the advance of epigenetics — which can be traced only through many overlapping stories — did not stop with DNA methylation and histone modifications. DNA was originally thought to be the determining party in a single, decisively important function: the production of RNA (via a process known as transcription) as an intermediate on the way to the production of protein (via translation of the RNA). However, certain small RNAs were discovered that did not, and could not, be translated into protein. They were found to be pursuing complex and varied pathways of their own, with equally complex and varied implications for the life of the organism, including its expression of its own genes.
For example, a given small RNA, in a complex with other factors, can target few or many protein-coding RNAs, leading to their degradation or otherwise impairing their translation into protein. The play of these degradative processes entails the give and take of numerous cellular activities, which is to say: not just DNA, but all these other processes as well determine how and under what circumstances a given DNA sequence will eventually yield a protein.
But things only get worse from here. There is also an activity known as RNA splicing, whereby the cell cuts up an RNA and then stitches together a selection of its pieces to make the final RNA that will be translated into protein. The process is almost unthinkably intricate, involving as it does hundreds of proteins plus various other molecules in a delicately coordinated play of forces. (How are we to think of the coordinating power? It’s not a question that gets asked very insistently.)
The ultimate result is that, depending on which pieces are stitched together, any one of a few or very many different protein variants might be produced — all originating from RNA derived from just one DNA sequence. Biologists have hardly begun to understand the reasons for the choice of protein variant, but it certainly has to do (at least) with the distinctive needs of different cell and tissue types. This, in turn, means that the larger cellular environment must somehow be able to express itself through all the coordinated intricacies of RNA splicing, directing the process toward one needed result or another.
Then there is RNA editing, in which the cell substitutes one “letter” of the RNA sequence for another, which is, in effect, to alter the original DNA sequence. And, short of this editing, there are wholesale chemical modifications of the RNA “letters”, which researchers have scarcely begun to investigate. On another front, molecular biologists are now looking into some of the thousands of long noncoding RNAs, which derive from the substantial portion of our DNA once dismissed as nonfunctional. These RNAs are now being found, one after another, to fulfill diverse roles in the biology of the cell, including many different forms of gene regulation.
And, again — perhaps most fundamentally — the fate of any given DNA sequence hinges on the dynamic, contextually induced “dance” of chromosomes in the three-dimensional space of the cell nucleus. (Yes, researchers sometimes find themselves drawn to this metaphor.) Gene expression is strongly shaped by the nuclear location of different chromosomal regions at any given time, and by the way the cell brings various chromosomal loci together for “socialization”. Or, as the head of a Harvard epigenetics laboratory puts it in rather different language:
It’s genomic origami ... In every cell, you have the same piece of paper. Stem cell, brain cell, liver cell, it’s all made from the same piece of paper. How you fold that paper determines if you get a paper airplane or duck. It’s the shape that you fold it into that matters. This has to be the 3-D code of biology.8
In other words (to shift the metaphor a bit, while avoiding the unjustified reference to “code”), the organism expresses something through its DNA at least partly in the manner a choreographer expresses something through the movements of a dancer. This is a truth of molecular biology that few have yet to reckon seriously with.9
We have hardly begun. But I suspect you have had enough.10 Do you understand epigenetics now?
Okay, a few remarks of a different sort may help.
The foregoing may suggest the existence of discreet epigenetic factors whose effects we need only add together in order to see the important thing, which is the net result in terms of gene expression. This is far from the truth. No single gene regulatory activity of the cell proceeds in isolation from all the others. The different activities are not in fact altogether different; they are thickly interwoven with each other and with the entire life of the cell. This is why molecular biologists find themselves looking more at patterns, networks, and contextual tendencies than at clear-cut causes. (More on causes and contexts in a moment.) No single type of molecule or structure is wholly definitive for any of the meaningful activity.
If the coordinated performance of more than three hundred proteins leads to the splicing of an RNA in such-and-such a way in this or that cell type, then those proteins are themselves subject to modifications that alter their functioning, and the enzymes that do the modifying are in turn influenced by still other molecules ... and there is no end to it. Further, some of the molecules most directly involved in splicing are directly regulated by the very splicing they help to regulate. So who is “controlling” whom?
Biologists knew that this sort of thing was going on from the very beginning, when the “DNA makes RNA and RNA makes protein” story was at its most naïve. For they knew that, before a cell could make protein from DNA, an enzyme — itself a protein — first had to transcribe DNA into RNA. Apart from transcription, there are numerous other ways proteins must manage — and modify and reshuffle — an organism’s DNA in order to preserve life. If protein depends on DNA, it is no less true that DNA depends on protein. But somehow it was easy to overlook the implications of such mutual entanglement, given the compelling causal simplicity and linearity of the governing story.
Today, of course, we know a great deal more about this transcribing enzyme. Its movement along a DNA sequence during transcription is more like a ballet performance than a chain of causes. Accompanied at the outset by a daunting corps of protein co-dancers, its progress is marked by intense molecular interactions and a rhythm of pauses that influences, among other things, what sort of RNA splicing will occur. Between the beginning and end of a gene sequence, the enzyme is subject to a changing combination of modifications (effected by other proteins) that enable its passage through this particular sequence with this particular result — a result that differs under the effect of different modifications and different dance partners.
The upshot of it all is that the statement we heard at the outset — “cells use their DNA code in different ways, depending on their jobs” — requires more than casual emphasis. “Using DNA in their own way” includes cutting and pasting, editing, and completely negating functional DNA sequences.
It’s not, in other words, just that the cellular orchestra “can perform one piece of music in many different ways”, as we read in the Nature news item. Rather, the organism can make many different pieces of music, as it does in the different cell types of the body, from bone to skin to muscle. Every organism certainly faces limits and constraints, often severe ones, but they should be seen as the limits and constraints of a coherent whole, not the iron dictatorship of a single molecule.
The Epigenome Roadmap papers hammer repeatedly on the themes of complexity and context-dependence. We hear,11 for example, of the “exquisitely tissue-specific epigenetic regulation” of thousands of long noncoding RNAs. We learn about “the value of interpreting DNA methylation and DNA accessibility in the context of integrated [chromosomal] states12”. We’re told that an intermediate form of DNA methylation is an “epigenomic signature of context-dependent function13”, and, again, that genomic variants bearing on autoimmune diseases appear to “exert highly contextual regulatory effects”.14
But there is in all this a great deal of confusion. Take, for example, the idea that, as cells differentiate, they undergo “context-specific rewiring” of various epigenetic factors, as one paper puts it.15 Where, we might wonder, is there any fixed and standard wiring waiting to be rewired if in fact the governing context is always to some degree fluid, dynamic, and shifting? Where do we ever see anything remotely analogous to wires constraining all the relevant molecules to go where they need to go, and to do so in the right time, in the right quantities, and with the right molecular partners?
The picture of a wired cell may sound conveniently causal, but it makes no sense. The confusion I just now referred to arises from conflict between the widespread recognition that organic contexts are an inescapable part of our understanding, on one hand, and an insistence on explaining things through the analysis of causal relations between separate parts, on the other.
What the references to context really imply — and what biologists seem constitutionally averse to recognizing — is that we’re dealing, at a biological level, with agency and intention in the organism. This agency is the constraining power, guaranteeing the peculiar sort of coherence we observe in organisms. The fact is inescapable. However little we may feel we understand it, it is written all over the literature of molecular biology. Moreover, the efforts to evade it result only in a worsening of the situation — which happens, for example, when, loath to attribute agency to the organism as a contextual whole, the researcher writes: “Transcription factors orchestrate the overall remodelling of the epigenome16”.
But this is absurd. Whatever might be said to orchestrate things in the cell cannot be just another thing in the cell, let alone one of the things that is so clearly being orchestrated. What’s really going on here is that material-minded molecular biologists, having been forced against their will to recognize in the organism a contextualizing (“orchestrating”) agency, immediately pivot and reassign this agency to particular molecules, as if such local, causally acting things could account for the global coordination of activities along a sustained storyline.
And so all the references to a governing context — and a context is not a context if it does not effectively establish, or govern, the relations among its parts17 — are conveniently eclipsed by what, in essence, are vitalistic molecules. Better to have vitalistic molecules — so the implicit thought seems to run — than any immaterial principle of coherence. At least the molecules have some sort of causal efficacy, even if such efficacy falls far short of any kind of “orchestration”. Isn’t that better than assigning causal agency to a “whole organism”? What can make things happen, if not the physical interaction of one part with another?
But the problems of biological causation and wholeness cannot be brushed away so easily. And it’s not as if the usual efforts to explain things by tracking down definitive causes and effects has ever looked promising.
A few years ago, in an article entitled “From Physical Causes to Organisms of Meaning”.18 I recited a litany of cautionary notes from molecular biologists about the chronic difficulties of disentangling cause and effect in epigenetics. The situation has not changed. “Even for the most studied epigenetic marks, only a correlation between [the marks and] active or repressed transcription is known”.19 Or, as the problem was summed up in Nature Reviews Genetics:
Knowledge of the interdependence of these marks remains sketchy; causality in terms of knowing what event triggers the next one is only understood for a small subset.20
The cognitive dissonance in that last statement could hardly be greater, since it is precisely the organic interdependence of the marks that makes the search for unambiguous causality fruitless, even for the supposed “small subset” of marks. Yes, so far as our knowledge of the interwoven epigenetic factors remains sketchy, we will certainly want to fill it out. We need to do so, however, not in order to tie down triggering causes and inevitable effects, but rather to gain a more detailed picture of the meaningful patterns through which the organism’s intentional agency is expressed.
Precisely this — the weaving together of interdependent causal factors by an organism pursuing its own aims — is what should tell us to abandon as a fools’ errand the search for a linear chain of causes and effects where one event “triggers” the next in a neat explanatory sequence.
Molecular biologists have proven remarkably resistant to any recognition of the way they confuse causal (“efficient”) and intentional explanation, invoking both in their descriptions while granting reality only to the former. Allow me to highlight the difference between the two — and the reality of biological intention in the organism — by making a large, but temporary, leap away from the molecular level.
Suppose I often go to the grocery store on Tuesday afternoons. How I get there may vary with the circumstances: I could walk, ride a bike, drive a car, hitchhike, take the bus. The path of the trip may also vary, depending, for example, on whether a road crew is doing work, or whether I come across a pedestrian who has just had a heart attack, or whether I encounter a friend who invites me to sit down for a cup of coffee for a few minutes. On one particularly eventful day, I might ride to the store in a hot air balloon. You will quickly realize that virtually every aspect of my grocery trip is subject to such variation.
There is no way we can generalize upon all the physical particulars, or their lawful aspects, so as to say, “These are the specific causes that account for a person’s going to the store”. Certainly something more or less like a causal account can be given in each different case. But what unifies and makes appropriate sense of the wildly disparate and contingent physical details is not itself a principle of physical science, materialistically conceived. Rather, the unifying explanatory principle lies in the fact that all the details bear witness to a consistent intention.
Anyone who wants to understand my life may be interested in tracing the various physically lawful aspects of my journeys to the store. But explanations at that level, shorn of intention as they are, can never adequately explain my going to the store. I go to the store because I intend to do so. Further, by observing all the physical particulars — not as explanations of what I do but rather as ever-varying constituents of a consistent intentional pattern — others can recognize my intentions and the way they are woven into the larger tapestry of my life.
Causes in the one sense — what we normally think of as physical causes — do not explain my going to the store in the second, intentional sense. Rather, in their infinite variability they reflect, or bear witness to, my intention. At the same time, my singular, unifying intention governs and makes sense of the diverse and never fully predictable physical activity.
Now, it is true that conscious intentions are a long way from cellular performances — although it’s worth remembering that my intentions somehow manage to embody themselves quite effectively in the muscle cells, blood flows, gene expression, and other physiological elements supporting my physical travel. But even considered in isolation, without a governing conscious intention, the molecular processes of the cell are recognized by every biologist as the bearers of intentions.
Molecules do not go to the store. But they are participants in activities such as cell division and DNA replication. If molecular biologists were not attending to these activities as reflections of cellular intention, and if they did not formulate research questions at least partly in terms of this intention, they would have no jobs. Their questions presuppose a kind of directed movement whereby a particular end is striven for despite the physical conditions never being the same as the last time the activity was carried out. The physical activities along the way to DNA replication no more explain the fact that the cell is undertaking this task than my activities on the way to the store explain my going to the store. In neither case is there a precise and reliable causal chain that equates to the intention, explains it away, or guarantees the end result. But the patterns of activity do bear witness to the task at hand.
In other words, we can watch and then know: this cell is setting about the work of cell division. We know in the way we get the sense of any storyline or narrative — a sense never reducible to the physical and causal relationships that incarnate the story. What we recognize are (among other things) intentions.
This is all the more true when we note how various disturbances — the death of a neighboring cell, an unusual ambient temperature, or the presence of some toxic molecules — may cause a “detour” on the way to cell division without necessarily derailing the overall process. Geneticists speak of cell cycle checkpoints, where a cell on a path toward division is prompted to “assess” its current physical state against its intentions and adjust its course accordingly. It may then, after a pause for course correction, continue on through the cell cycle, or else arrest the cycle and forgo division, or else, if the troubles are too threatening, set in motion an orderly cell death.
In the case of cells that continue on through division, it is obvious enough that the nature of the different possible threats leading to the checkpoint will shape the cell’s corrective behavior, leading to very different cellular trajectories. But even a single type of problem can result in different pathways through the trouble. One of the conditions producing a checkpoint pause is DNA damage caused, for example, by ultraviolet solar radiation or reactive oxygen species in the cell. It turns out that the same or similar damage leads to different repair responses, depending, among other things, on the current stage of the journey toward cell division. That is, of the various different (and extraordinarily elaborate) processes of DNA repair a cell can undertake, the one employed in a particular circumstance hinges on the current cell cycle context.
Dismantling the DNA damage checkpoint
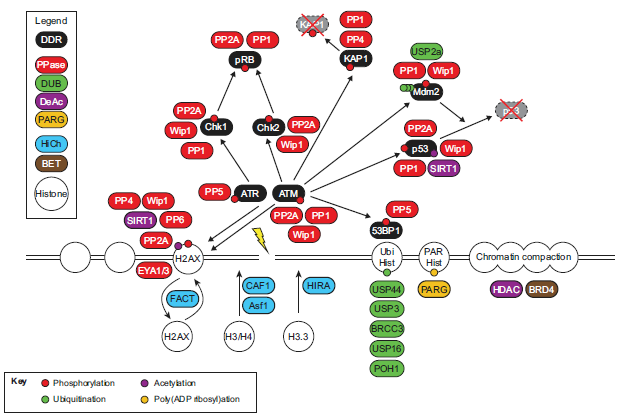
This figure, taken from Shaltiel et al. (2015),* shows some of the enzymes and other molecules required for undoing molecular modifications previously made in support of DNA repair activities. The authors summarize part of the story this way (don’t worry about the technical details!):
“Activation of the DNA damage response leads to a highly complex series of events orchestrated by a plethora of [protein] posttranslational modifications, such as phosphorylation, acetylation, methylation, poly(ADP-ribosyl)ation (PARylation), ubiquitylation and small ubiquitin-like modifier (SUMO)ylation. As these modifications trigger checkpoint activation, cell cycle arrest and DNA repair, removal of these modifications by dedicated enzymes or degradation of modified proteins is essential for checkpoint silencing and recovery. Thus far, this emerging field has identified redundancy and complex cross-talk between the modifications as a common theme”.
As the authors indicate here, undoing the chemical modifications intended to serve the processes of DNA repair is not the only option. The cell may also simply remove a protein by degrading it and freeing its constituents.
Likewise, the way in which the DNA damage response is eventually silenced — and it must be silenced in time in order to prevent unnecessary cell death — differs depending on the phase of the cell cycle.21 Keep in mind that undoing the almost incomprehensible variety of molecular changes required for the DNA damage response is fully as complex as the initial activation of the response. (See accompanying box.)
It is impossible even to imagine that, on any two occasions, all the reactions and interactions will take place in exactly the same way, with exactly the same results, leaving the cell in exactly the same state. One can even remove altogether some of the molecules shown in the figure, and yet the overall process proceeds more or less normally — which is one reason why the authors of the illustration refer to the redundancy and the cross-talk through which the cell’s options and possible pathways are diversified.
It’s long been known that causality in organisms manifests through countless factors in a complex, mutual embrace, where none of those factors can be isolated from the others as an unambiguous cause of any significant state of the organism. There’s good reason why molecular biologists fondly refer to diagrams of protein interactions as “hairballs”. From the zygote onward the organism is an intentional whole determining its own parts, not a passive resultant determined by its parts. This point has been acknowledged throughout the past 250 years or so by many of those biologists and philosophers who have paused to think about the matter.
That molecular biologists should simply ignore this understanding and, as a result, entangle themselves in hopelessly contradictory terminology — that they should make “orchestrating” molecules intentional while denying intention where we actually see it, in the organism as a whole — well, it’s not easy to fathom. But it surely has something to do with the reigning taboo among biologists against actually believing their own pervasive and discipline-wide characterizations of the organism as an intentional, and therefore in some root biological sense “mental” or “thoughtful”, agent.22
The good news about the Epigenome Roadmap papers is that, despite the obvious persistence of old mindsets, researchers have in some regards moved forward. In particular, the realities of the organism appear to have persuaded at least some of them that the ambiguities of cause and effect need not interfere with their efforts to make sense of significant pattern, or form. This sort of effort seems to be the main business of the various authors, and the cumulative effect of the pictures they draw is compelling.
But those pictures also suggest that the term “epigenetics” is of minimal usefulness. The word testifies to an old and tired conviction that DNA is what really counts, and all else is a mere add-on. This fantasy has no place in today’s biological science. Nor are there distinct “layers” of gene regulation, one or several of which we can cleanly isolate as “epigenetic”. Whenever we look at particular activities characteristic of a given type of cell, we see “complex regulatory processes in which ‘epigenetic’ and ‘nonepigenetic’ components are entwined”.23
This is not to suggest that organic activities lack structure and differentiation, or that particular functions are not more or less associated with one or another part of the organism. Organisms are, it sometimes seems, almost miraculously structured. But they possess no self-contained “modules” (to use a mechanical or computational term many biologists are fond of) unreceptive to the interpenetrating and mutually defining influences constituting the life of the organism.24
Biologists are in fact discovering more and more evidence for what they term “crosstalk” between activities once considered to be functionally self-contained. It is becoming increasingly hard to find molecules connected with one process that do not also have connections with other processes initially thought to be unrelated to the first. The “surprising” multifunctionality of molecules in the cell is one of the familiar themes of the contemporary literature.25
To my mind, the most obvious value of the current work on “epigenetics” lies in the way it highlights the centrality of organic and dynamic context for biological understanding, which is much the same as highlighting the centrality of intention and agency. But nearly all biologists today remain dead set against any recognition of intention, agency, or any other “interior” dimension of the organism. They have been raised and trained in an implicit and severe dualism, which is the prerequisite for thinking it possible to dismiss half the falsely dichotomized world out of hand. The aspects of reality they want to dismiss had first to be separated by violence from the aspects they want to keep.
But now I risk venturing far beyond epigenetics, and spitting out more words than you signed on for at the beginning. Better to stop here.26
Notes
2. The historical derivation of epigenetics is actually rather less straightforward than is sometimes suggested today. Its origin in 1942 (see Waddington 2012*) traces in part back to the much older term epigenesis, which referred to the becoming, unfolding, or development of a mature organism from its germ — all without particular reference to genes or the modern discipline of genetics. The reference to genetics, however, was already present alongside epigenesis in Waddington’s usage, and it came out ever more strongly with the passage of time, so that we might reasonably think of today’s epigenetics as constructed solely upon genetic in the biomolecular sense rather than genesis in the developmental sense. But gene and genetic themselves involve complexities, which I cannot go into here. For a survey of the thought leading from Aristotle to the modern concept of the gene, see chapter 1, “Genesis of the Gene”, in Lenny Moss’ What Genes Can’t Do (2003*).
3. The reader will note that I have said nothing about the inheritance of epigenetic features. This is best reserved for my concluding footnote.
4. See my article “Biology’s Shameful Refusal to Disown the Machine-Organism” (Talbott 2014a*).
6. Of course, whatever has the power to alter the functions of DNA might be thought to take precedence over DNA. Fancy footwork is always required of those biologists who acknowledge such powers while still wanting to preserve their faith in genes as First Causes. It is difficult — so writes evolutionary and developmental biologist Scott Gilbert — to lay hold of the “tertium quid” (third something) that “generate[s] the phenotype from genotypic guidance” (Gilbert 2012*). Difficult indeed, when, having recognized the organism’s power to employ its physical resources (including genetic resources) in “generating” its own developing form, you refuse to give up the notion of “genotypic guidance” — that is, the notion that it is really the genes that make the essential choices leading to that form.
The only way to resolve the contradiction between “employing DNA”, on the one hand, and “being caused or guided by DNA”, on the other, is by jettisoning notions of one-sided causation by a particular part of the organism. Then we can recognize that the “third something” producing the phenotype is not actually a third thing lying between genotype and phenotype; it is the whole organism, as we will see below.
7. See, for example, Ptashne, Hobert and Davidson (2010)*. In complaining about the project of the International Human Epigenome Consortium, the authors refer to a similarly critical 2008 letter signed by “eight prominent scientists” and available at https://madhanilab.ucsf.edu/epigenomics. However, as of this writing the letter has been removed from that location. One hopes that good sense has prevailed and that at least some of the signatories may no longer feel comfortable with the letter’s badly outdated criticisms. An abbreviated form of the letter was published in Science (Madhani et al. 2008*).
8. John Rinn, Associate Professor in the Department of Stem Cell and Regenerative Biology at Harvard University and Medical School, and Senior Associate Member of the Broad Institute. Quoted in Zimmer 2015*. I should add that it is not the “same piece of paper”; we do not find any chromosome or stretch of DNA to be the same in different cell types. There is a continual and substantive transformation going on, as mentioned above in the discussion of epigenetic marks. Even the constellation of water molecules around the double helix affects what it is and how it functions (Talbott 2013*).
9. The obstacle to any serious reckoning is the prevailing conviction that we are merely looking at molecular mechanisms. But there are no such mechanisms in the cell. There are lawful performances, to be sure, but these are radically different from the various sorts of devices we humans contrive. For much more on this theme, see the articles listed here.
10. Anyone who wants an overview of the scores of topics we could pick up here — many of which would require book-length treatment if justice were to be done to their complexity — may want to glance through just the outline of contents in the collection of (very informal) notes, “How the Organism Decides What to Make of Its Genes”.
11. Amin, Harris, Onuchic et al. 2015*.
12. Roadmap Epigenomics Consortium 2015*.
13. Elliott, Hong, Xing et al. 2015*.
14. Farh, Marson, Zhu et al. 2015*
15. Tsankov, Gu, Akopian et al. 2015*.
16. Tsankov, Gu, Akopian et al. 2015*.
17. It’s important to realize that a context is never a “mindlessly” material thing. To use an example I’ve offered before: a chess board and the associated game pieces, considered merely as an aggregation of things, are not a context. They are just “stuff”. What makes them part of a context, giving them meaning, are the rules of the game, the relationship and intention of the players, the cultural setting for the game, and so on. These immaterial realities are what constrain an infinitely variable set of physical activities, shaping them to the form of the game.
Of course, in the case of chess we require (at least initially) an exercise of conscious attention and choice in order to adhere to the form of the game. But it need not always be so. A capable pianist might improvise upon a theme while consciously unaware of the “rules” of melodic, harmonic, and aesthetic relationship he is respecting. And I have tried to show (in “Psyche, Soma, and the Unity of Gesture”) that we have no grounds for radically separating our conscious intentions from the intentions we can recognize at work in our bodies, all the way down to the cellular and molecular level.
“Context” is one of the many terms molecular biologists rely on without giving any disciplined thought to what must be implied by the term if it is to carry the meanings they require of it.
20. Lenhard, Sandelin and Carninci 2012a*.
24. In the nineteenth century Samuel Taylor Coleridge offered what seems to me an excellent way to conceive the relation between parts and whole in organisms of different complexity: “The living power will be most intense in that individual which, as a whole, has the greatest number of integral parts presupposed in it; when, moreover, these integral parts, together with a proportional increase of their interdependence, as parts, have themselves most the character of wholes in the sphere occupied by them” (quoted in Barfield 1971*). The human heart is an impressive whole considered in and of itself. Yet it cannot in the end be considered adequately in that way, since the extraordinary degree of its own perfection is matched by the degree of its dependence upon, or interwovenness with, the rest of the body. This mutual dependency is much greater than we find when examining the circulatory system of simpler, relatively undifferentiated organisms.
The concept of polarity is valuable here. A bar of iron may be unmagnetized or weakly or powerfully magnetized. It remains a unity in all cases, but the more intense its magnetization — the more its north pole is differentiated from its south pole — the stronger the forces holding the two poles in unity turn out to be. Similarly in the organism: the more highly differentiated and perfected the individual organ becomes, the more elaborate are its means of contextual integration. And, I might add, the same holds true of society: the strongest, most independent and centered individual is the one who has most healthily internalized the needs of the larger society, so that pursuing her own necessities and serving society’s needs are not two separate things.
25. Of course, as we learn more and more about this multifunctionality — which is to say, as we learn about the interconnectedness and unity of the organism — we have ever less reason to think that the “unrelated” processes are in fact unrelated. So, with time, the surprising results look ever less surprising; mentioning them provokes only a yawn. That’s progress.
26. Or not quite stop, if I can tie up what some readers may see as a loose end. At the outset of this article I suggested a broad definition of epigenetic, saying that it concerns “all the ways DNA is caught up in the activity of its larger context and brought into service of the whole”. I thereby omitted any reference to the heritability of epigenetic factors. This was intentional. Heritability is part of the conventional definition because DNA remains central to conventional thinking, and DNA has long been equated (disastrously) with what is heritable. So the bar was set accordingly: “If you want your epigenetic effects to be taken seriously, you must show that they are heritable”.
Heritable is usually applied to “stably heritable features” — fixed structures capable of being passed down through a continuous series of reproductive cycles. The idea is that if a change is to have real significance, it ought to be more or less indefinitely sustainable in the form of some recurring physical entity. But there is no justification for making this requirement absolute — certainly not at the cellular level where, at least during differentiation, if not more subtly at all times, cells require coherent and successive change in order to pursue the destinies of their particular lineages. Each cellular generation carries further what the previous one achieved, adding change to change rather than preserving everything unchanged. It is the fluent sequence of changes that needs to be sustained if cell differentiation is to proceed successfully. The organism as a whole is clearly capable of directing this process.
As for evolution: despite the still-dominant role of population genetics in evolutionary thinking, there is no reason for the theorist to focus solely on the stable maintenance of mutations in genetic (or epigenetic) structures as opposed to processes of more or less continuous and consistent change. What makes the more fluid notion of inheritance seem crazy to evolutionary biologists is their inability to conceive organisms, singly and collectively, as active and capable agents of evolutionary change rather than as passive bearers of atomistic genes mutating randomly. We do, however, see the barest hint of a more adequate conceptualization of the evolving organism in discussions of the evolution of robustness and evolvability. (But I truly mean barest. There is no sign yet of escape from the oppressive materialism that has for so long prevented explicit acknowledgment of the organism as an intentional agent.)
Furthermore, in the case of both differentiating cells and evolving organisms, current research techniques hardly put us in a position to pronounce upon the stability or instability of changes we observe — not when those changes are found to inhere in patterns of activity rather than fixed structures. We can, however, hope for the development of more adequate techniques, since molecular biologists are increasingly being forced to shift their attention away from fixed structures and toward the live patterns found in plastic pathways and functions, dynamic networks, and adaptive systems. But this work will never bear the promised fruit so long as the scientist avoids considering these live patterns in their own intentional terms, and instead tries to explain them by adverting solely to things and causes.
All this, of course, is badly in need of more leisurely elaboration, which I hope to offer before long.
Sources:
Amin, Viren, R. Alan Harris, Vitor Onuchic et al. (2015). “Epigenomic Footprints Across 111 Reference Epigenomes Reveal Tissue-Specific Epigenetic Regulation of lincRNAs”, Nature Communications (online publication: Feb. 8). doi:10.1038/ncomms7370
Barfield, Owen (1971). What Coleridge Thought. Middletown CT: Wesleyan University Press.
Elliott, GiNell, Chibo Hong, Xiaoyum Xing et al. (2015). “Intermediate DNA Methylation Is a Conserved Signature of Genome Regulation”, Nature Communications (online publication: Feb. 18). doi:10.1038/ncomms7363
Farh, Kyle Kai-How, Alexander Marson, Jiang Zhu et al. (2015). “Genetic and Epigenetic Fine Mapping of Causal Autoimmune Disease Variants”, Nature vol. 518 (Feb. 19), pp. 337-43. doi:10.1038/nature13835
Felsenfeld, Gary (2014). “A Brief History of Epigenetics”, Cold Spring Harbor Perspectives in Biology vol. 6, no. 1 (Jan.). doi:10.1101/cshperspect.a018200
Gilbert, Scott F. (2012). “‘The Epigenotype’ by C. H. Waddington”, International Journal of Epidemiology vol. 41, pp. 20-3. doi:10.1093/ije/dyr186
Lenhard, Boris, Albin Sandelin and Piero Carninci (2012a). “Metazoan Promoters: Emerging Characteristics and Insights into Transcriptional Regulation”, Nature Reviews Genetics vol. 13 (April), pp. 233-45. doi:10.1038/nrg3163
Madhani, Hiten D., Nicole J. Francis, Robert E. Kingston et al. (2008). “Epigenomics: A Roadmap, But to Where?” letter available at https://madhanilab.ucsf.edu/epigenomics and abbreviated in Science vol. 322, pp. 43-4. doi:10.1126/science.322.5898.43b
Moss, Lenny (2003). What Genes Can’t Do. Cambridge MA: MIT Press.
Ptashne, Mark, Oliver Hobert and Eric Davidson (2010). “Questions over Scientific Basis of Epigenome Project”, Nature vol. 464 (March 25), p. 487. doi:10.1038/464487c
Roadmap Epigenomics Consortium (2015). “Integrative Analysis of 111 Reference Human Epigenomes”, Nature vol. 518 (Feb. 19), pp. 317-29. doi:10.1038/nature14248
Roy, Ananda L. and Dinah S. Singer (2015). “Core Promoters in Transcription: Old Problem, New Insights”, Trends in Biochemical Sciences vol. 40, no. 3 (Mar.), pp. 165-71. doi:10.1016/j.tibs.2015.01.007
Shaltiel, Indra A., Lenno Krenning, Wytse Bruinsma and René H. Medema (2015a). “The Same, Only Different — DNA Damage Checkpoints and Their Reversal Throughout the Cell Cycle”, Journal of Cell Science vol. 128, pp. 607-20. doi:10.1242/jcs.163766
Smith, Kerri (2015). “Epigenome: The Symphony in Your Cells”, Nature (online publication: Feb. 18). doi:10.1038/nature.2015.16955
Talbott, Stephen L. (2010). “Reframing the Mind-Body Problem: An Exercise in Letting Go of Dualist Assumptions”. Available at http://BiologyWorthyofLife.org/mqual/epist.htm
Talbott, Stephen L. (2011b). “From Physical Causes to Organisms of Meaning”, The New Atlantis no. 30 (winter), pp. 24-49. Original version published as “What Do Organisms Mean?” in NetFuture #182 (Feb. 22). Available at http://BiologyWorthyofLife.org/mqual/genome_6.htm
Talbott, Stephen L. (2013). “The Unexpected Phases of Life”. Available at http://BiologyWorthyofLife.org/org/comm/ar/2013/phases-of-life_13.htm
Talbott, Stephen L. (2014a). “Biology’s Shameful Refusal to Disown the Machine-Organism”. Available at http://BiologyWorthyofLife.org/org/comm/ar/2014/machines_18.htm
Talbott, Stephen L. (2014b). “How Does an Organism Get Its Shape: The Causal Role of Biological Form”. Available at http://BiologyWorthyofLife.org/org/comm/ar/2014/brady_24.htm
Talbott, Stephen L. (2014c). “Psyche, Soma, and the Unity of Gesture” (Part 2 of “From Bodily Wisdom to the Knowing Self”). Available at http://BiologyWorthyofLife.org/comm/ar/2014/bodily-wisdom2_20.htm
Talbott, Stephen L. (2014d). “Let’s Loosen Up Biological Thinking!”. Available at http://BiologyWorthyofLife.org/org/comm/ar/2014/mental_cell_23.htm
Tsankov, Alexander M., Hongcang Gu, Veronika Akopian et al. (2015). Nature vol. 518 (Feb. 19), pp. 344-9. doi:10.1038/nature14233
Waddington, C. H. (2012). “The Epigenotype”, International Journal of Epidemiology vol. 41, pp. 10-13. doi:10.1093/ije/dyr184. Reprint of “The Epigenotype”, Endeavour (1942), pp. 18-20.
Zimmer, Carl (2015). “Is Most of Our DNA Garbage?” New York Times (Mar. 5). Available at https://nytimes.com/2015/03/08/magazine/is-most-of-our-dna-garbage.html
This document: https://bwo.life/org/comm/ar/2015/unthinking_epigenetics_27.htm
Steve Talbott :: How to Unthink Epigenetics

